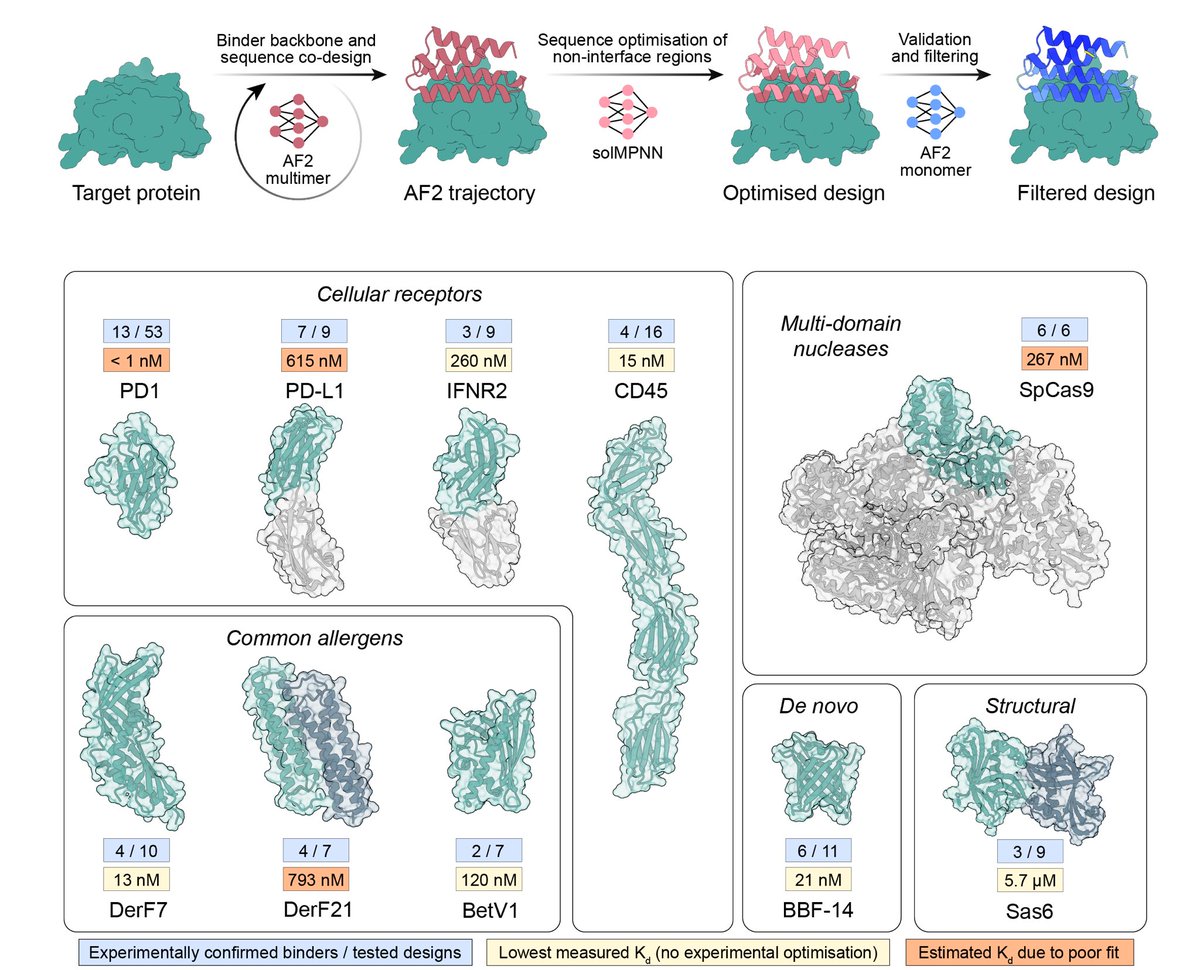
I decided to release a couple of figures from my PhD thesis, perhaps they might be useful to someone! First off is a remake of the famous Makarova et al. 2020 CRISPR classification. This one also includes a schematic of the targeted nucleic acid. 



Next off we have structural renderings and domain schematics of Class 2 effectors released at the time of writing (missed Cas12k then). PDBs: 6O0Z, 6I1K, 5U30, 6NY2, 7C7L, 6XMG, 6W5C, 5XWP 



Then we get onto the focus protein - SpCas9.First we have structural renderings of the apo, binary state and the guide RNA. PDBs: 4CMP, 4ZT0 

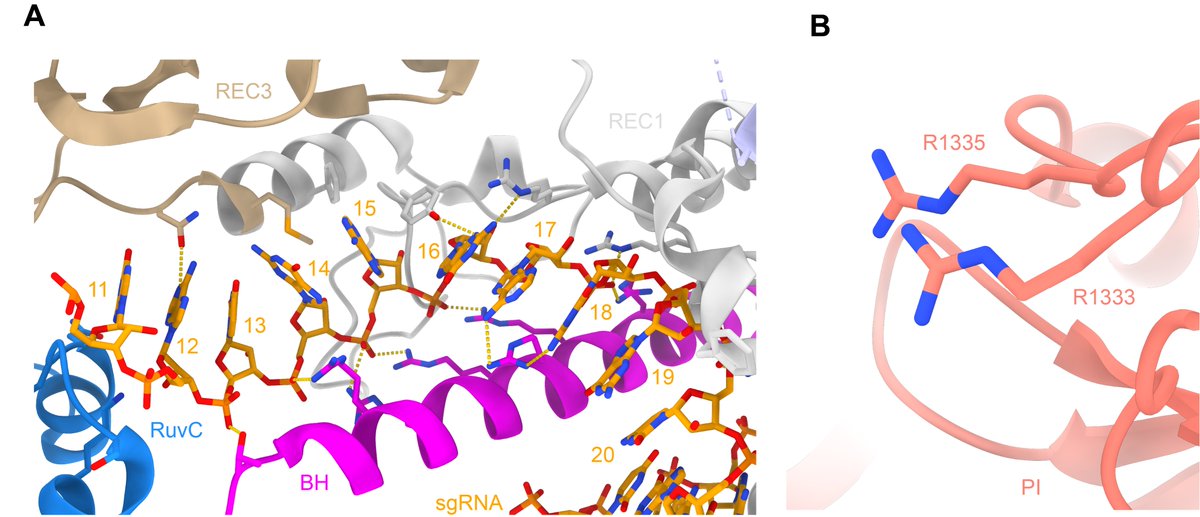
Here I highlight the structural preordering of the guide RNA seed region in a pseudohelical arrangement (A) and the PAM-interacting residues in the binary complex. Both of these significantly increase the efficiency of target sampling and binding. PDB: 4ZT0 
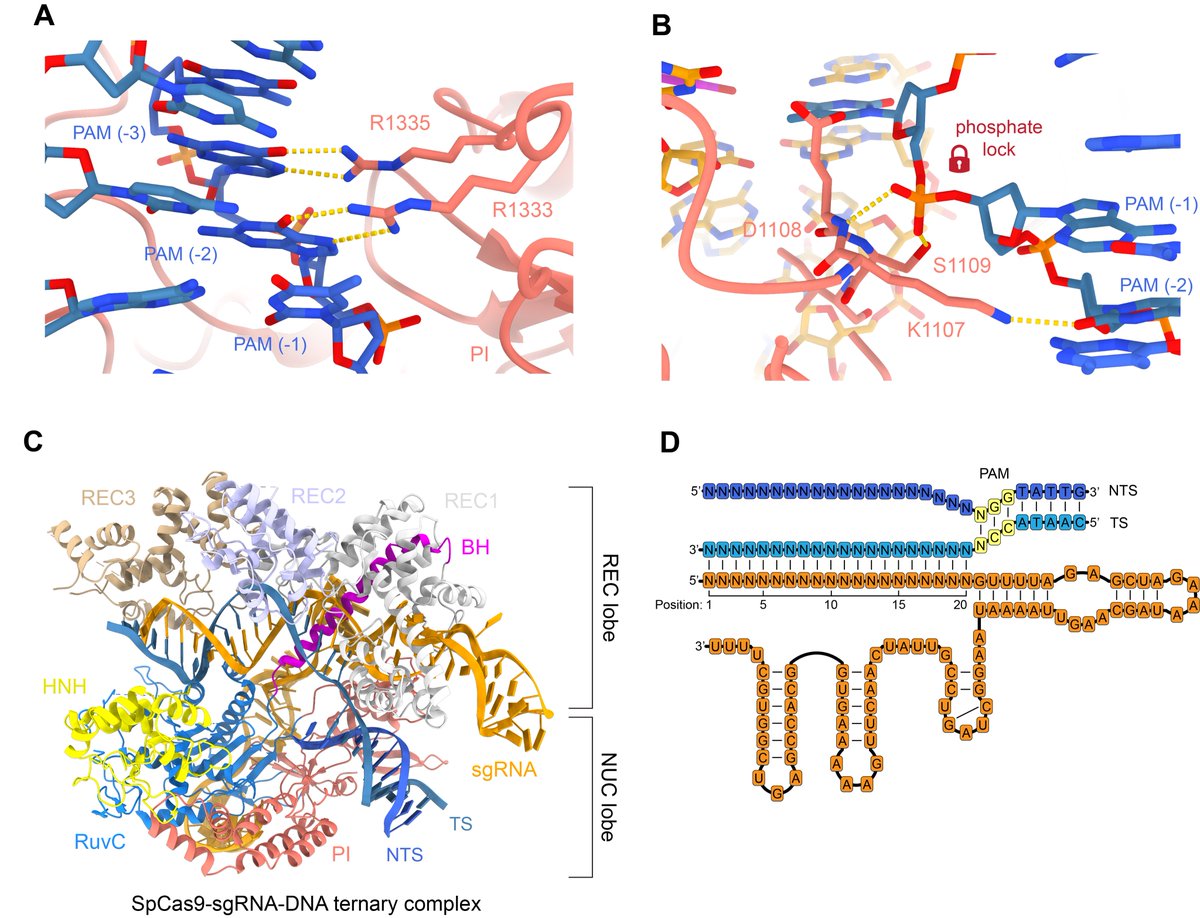
Once the DNA target is bound, the PAM-interacting arginines specifically recognise two GG nucleotides (A). The PAM-proximal phosphate is stabilised (B) to allow base pairing with the guide RNA. PDBs: 4UN3, 6O0Z 
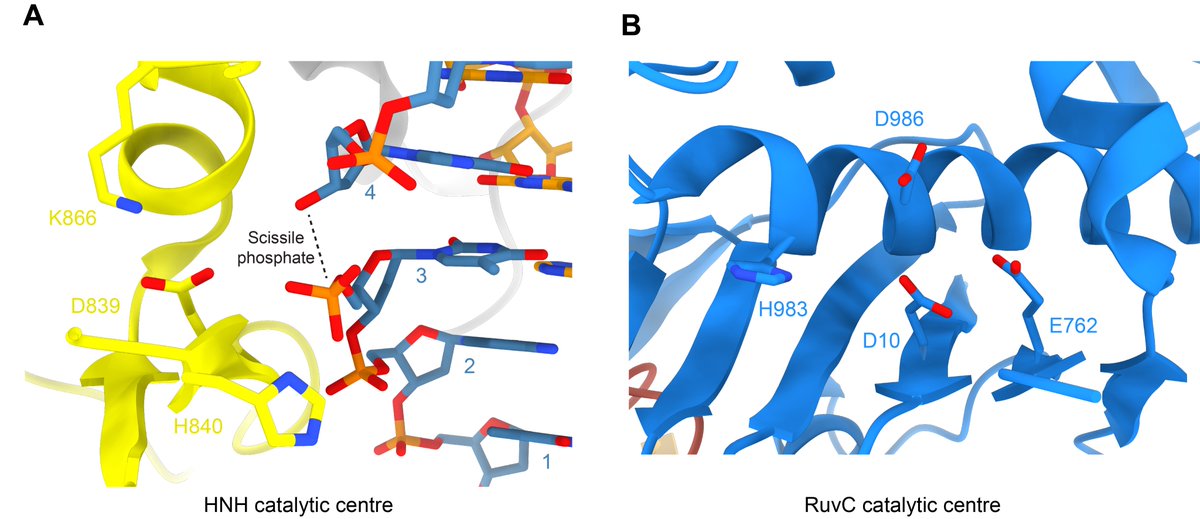
Lastly we have the renderings of the HNH and RuvC nuclease sites and active residues. PDBs: 6O0Y, 4UN3 
• • •
Missing some Tweet in this thread? You can try to
force a refresh