
Very happy to see our microRNA metastases paper out. co-led by @HyeEirik and by many fantastic colleagues
We show the reasons for previously published contradicting reports of de-regulated #microRNAs in colorectal cancer #tumors and #metastases:
tinyurl.com/yckwdk2z
We show the reasons for previously published contradicting reports of de-regulated #microRNAs in colorectal cancer #tumors and #metastases:
tinyurl.com/yckwdk2z
Among them are
- lack of correction for background expression
- different metastatic sites were not analyzed as separate entities
- expression cut-off level for microRNAs to ensure biological relevance are typically not used
- lack of correction for background expression
- different metastatic sites were not analyzed as separate entities
- expression cut-off level for microRNAs to ensure biological relevance are typically not used
We establish a stringent, reproducible and easily expandable bioinformatics pipeline (github.com/eirikhoye/mirn…) that we applied on all previously published (depending on availability) and a large range of novel smallRNA sequencing datasets, altogether 268 datasets.
We find that microRNA expression of correctly performed experiments is independent of study and resemble location in the body (i.e. organ) academic.oup.com/view-large/fig… 
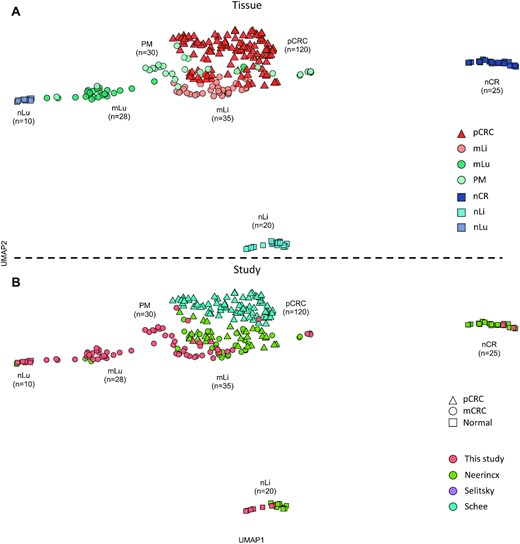
metastatic sites, while sharing some deregulated miRNAs, differ in microRNA expression academic.oup.com/view-large/fig… 

We also show that cell-type specific microRNAs inform about cellular heterogeneity in tissues, rather than deregulation academic.oup.com/view-large/fig… 

Finally, we show a set of clearly deregulated microRNAs in metastases with strong potential as biomarkers and for future mechanistic studies. academic.oup.com/narcancer/arti… 

This work was heavily inspired by previous work from @Marc_Halushka @Mendell_lab @KennethWWitwer using the tools of the @FriedlanderLab and others, not on Twitter.
Thank you for comments and help with data acquisition @MichaelHackenbe
Thank you for comments and help with data acquisition @MichaelHackenbe
This study started back in 2014 and actually made us realize, back in 2015, that the human microRNA complement needed reannotation. which ultimately led to the creation of @MirGeneDB
If you are interested in our approach see @HyeEirik presenting part of the approach and results here
during our smallRNA bioinformatics club series (smallrna-bioinformatics.eu)
during our smallRNA bioinformatics club series (smallrna-bioinformatics.eu)
@HyeEirik @threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh




