As Omicron has washed over the US, infecting perhaps 25% of the population already & likely to reach 40% by mid-February—see thread by @trvrb below—it has driven down almost all other respiratory pathogens, with one curious exception I’ll get to later. 1/9 
https://twitter.com/trvrb/status/1483996723458445319

This is not entirely unexpected. Viral infections trigger both innate and adaptive immune responses that can prevent infection by other viruses. Behavior changes likely contribute to this pattern as well. 2/9
https://twitter.com/pjie2/status/1470430087510175755
There have been some claims that rhinovirus infection protects against SARS-CoV-2 infection. As you can see in the graph below, SARS-CoV-2 and RV prevalence seem almost perfectly inversely related in recent months. 3/9 news.yale.edu/2021/06/15/com… 
https://twitter.com/MackayIM/status/1380135766257397763

Rhinoviruses typically peak in spring & fall & are lower during winter, when influenza, coronaviruses, HMPV, RSV, & other viruses thrive, suggesting these viruses & RVs compete & inhibit one another in some way. Great article on RVs by @MackayIM below. 4/9 virologydownunder.com/rhinovirus-ram…
One would expect more closely related viruses to compete most vigorously. The rapid displacement of one SARS-CoV-2 variant by another in this pandemic seems to confirm this. 5/9 

The history of influenza also suggests closely related viruses undergo intense competition. It’s thought H3N8 dominated in late 19th c. but was replaced by H1N1 in the 1918 pandemic. H1N1 in turn was displaced by H2N2 in 1957, which vanished when H3N2 appeared in 1968. 6/9 

Curiously, H3N2 has persisted through the emergence of H1N1 in 1977 (possible lab leak) & pH1N1 in 2009. The paper linked to below suggests this is because the 2 major proteins on their surfaces are quite different & antigenically distinct. 7/9 journals.asm.org/doi/10.1128/mB… 6/ 



This makes the one exception to the downward trend in all non-Omicron respiratory pathogens all the more remarkable. As Omicron has risen, everything else has fallen—except the seasonal coronaviruses, which have risen in step with Omicron. 8/9 
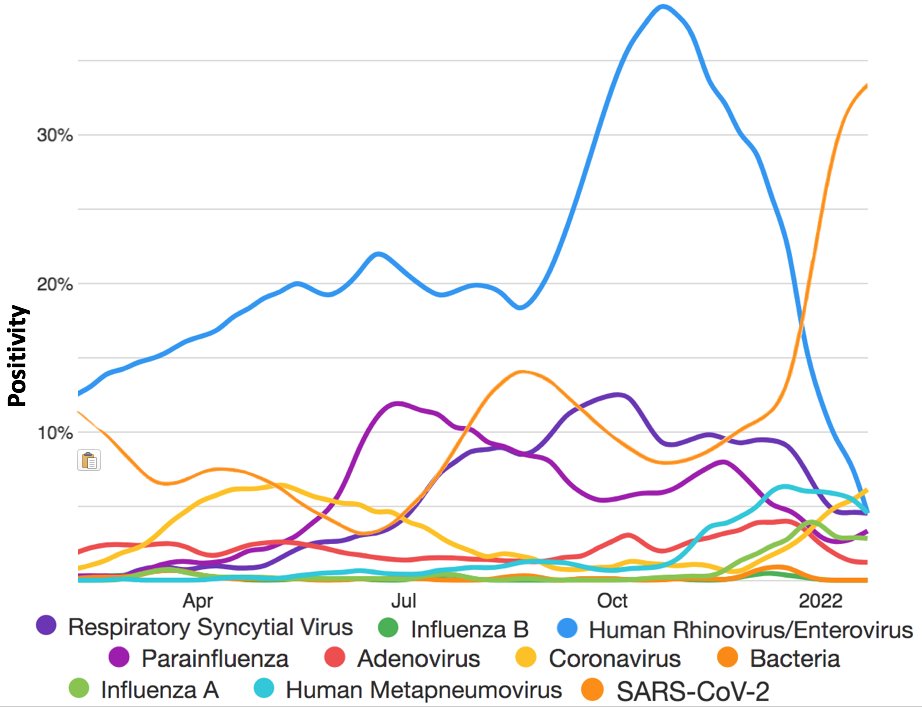
OC43, NL63, and 229E have all contributed to the recent rise in seasonal coronavirus cases. (HKU1 has been absent for nearly 2 years now.) I don’t have any idea why this would happen. I’d love to hear what others’ hypotheses are though. 9/9 

• • •
Missing some Tweet in this thread? You can try to
force a refresh