Rudi Jaenisch was harassed by the woke mob for publishing a paper that could incite "vaccine hesitancy".
Looks like his hesitancy was was in order.
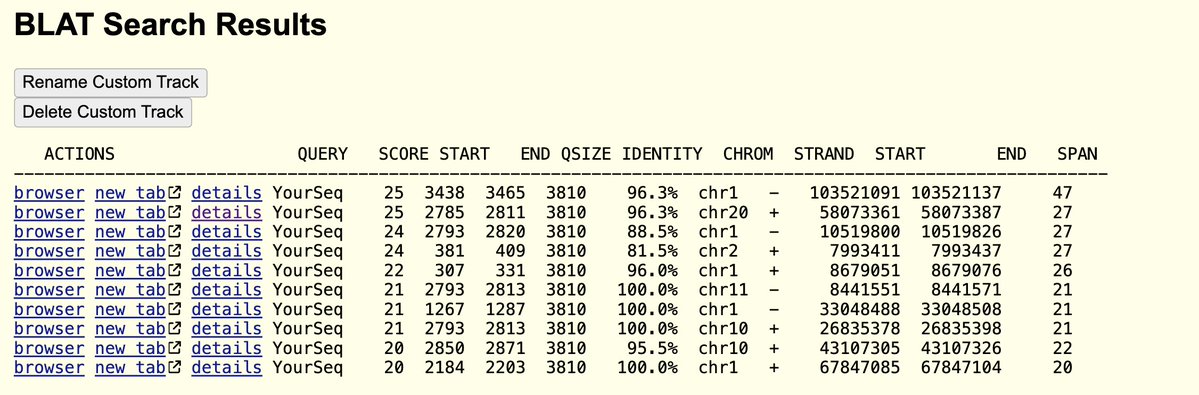
This paper demonstrates the BNT162b2 mRNA is getting reverse transcribed into DNA. It DOES NOT show
google.com/url?sa=t&rct=j…
Looks like his hesitancy was was in order.
This paper demonstrates the BNT162b2 mRNA is getting reverse transcribed into DNA. It DOES NOT show
google.com/url?sa=t&rct=j…
Integration into the genome... yet. They need long read Whole genome sequencing to prove that which Im sure is currently underway. Rudi showed it was integrated but was critiqued that it could be an artifact of making sequencing libraries. This paper fills in many questions. 
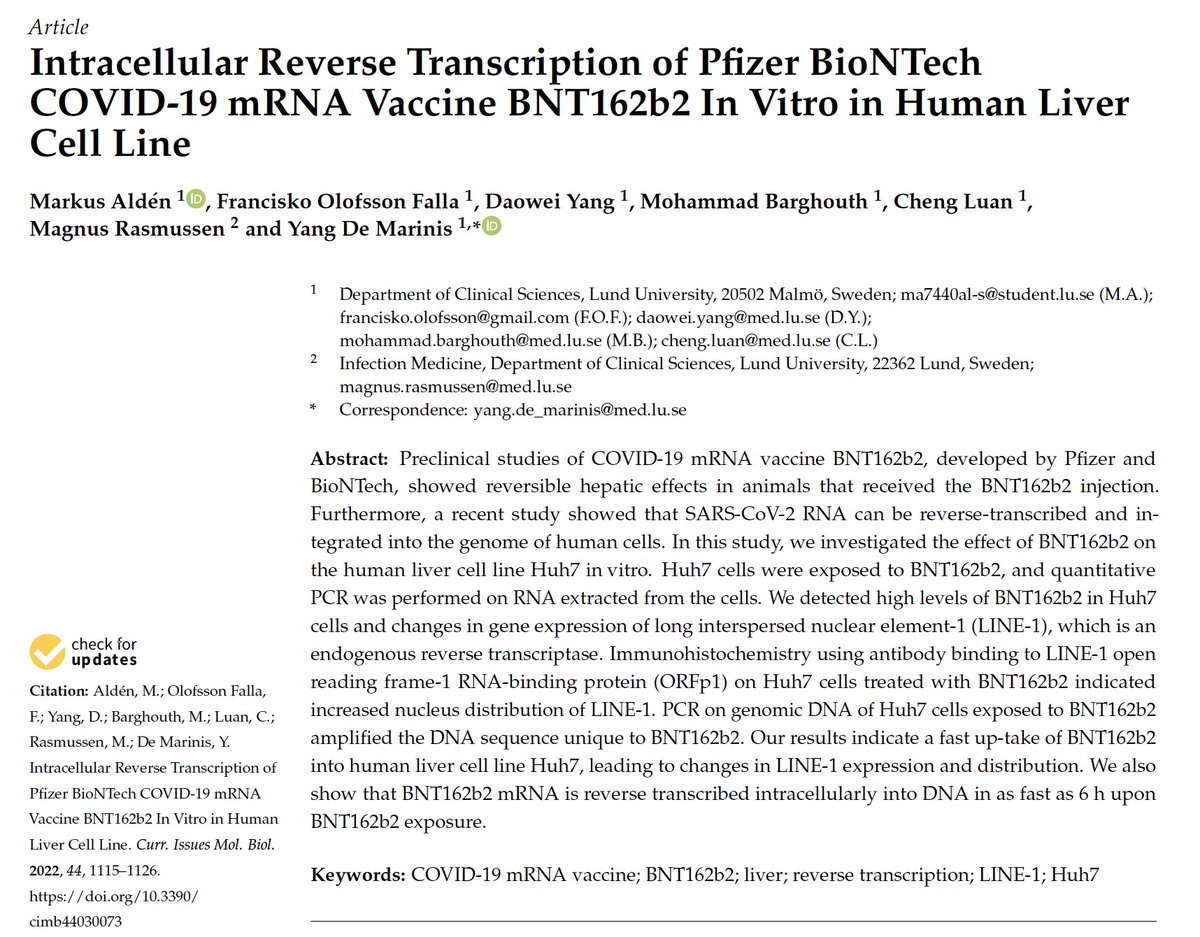
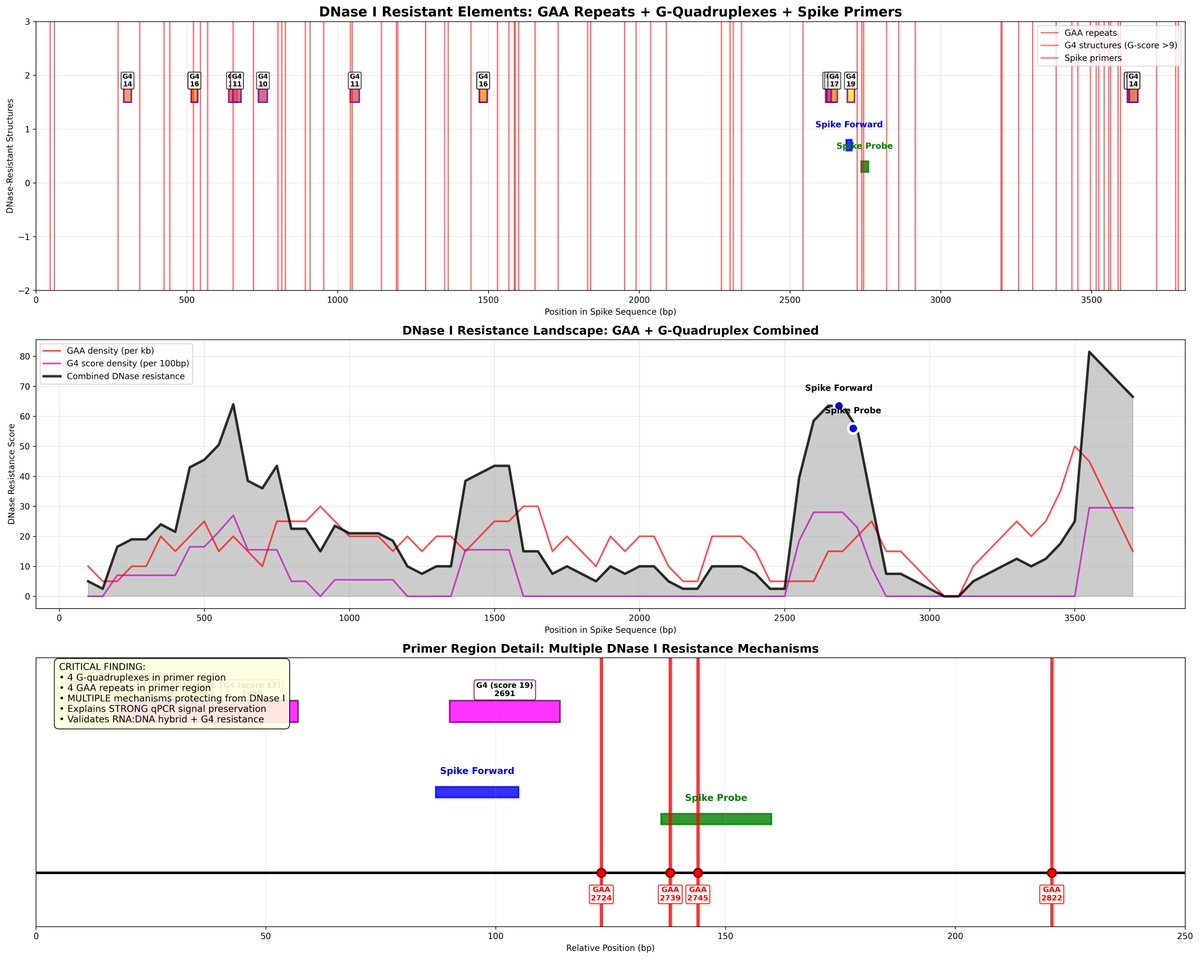
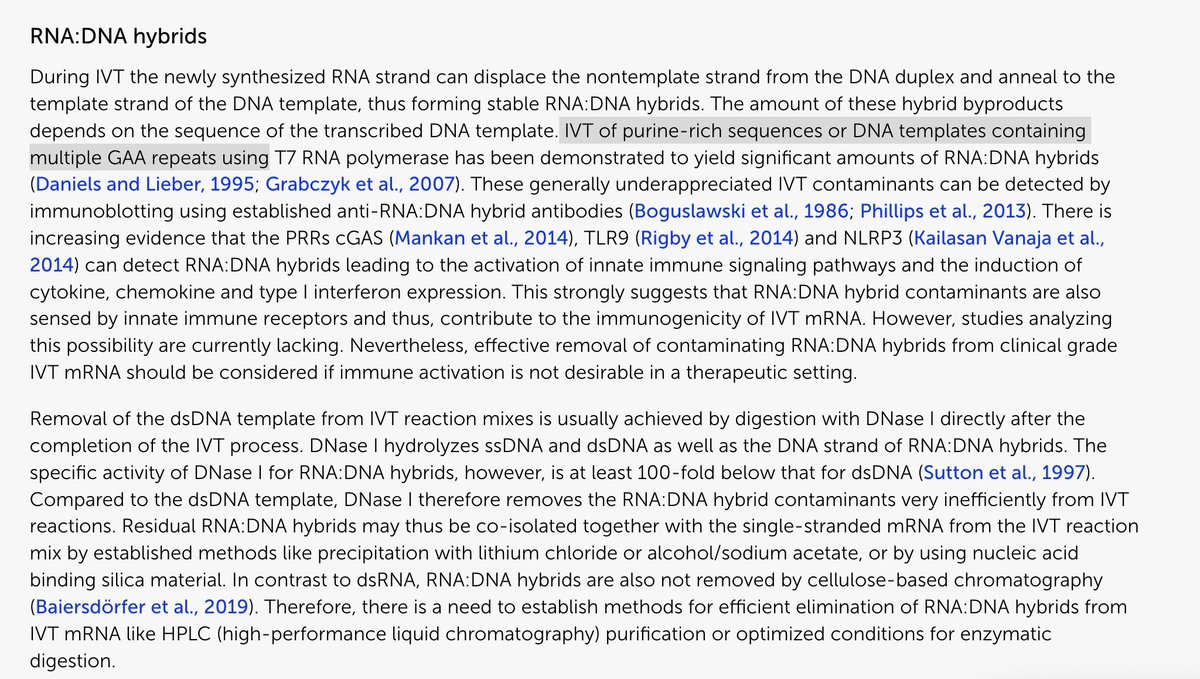
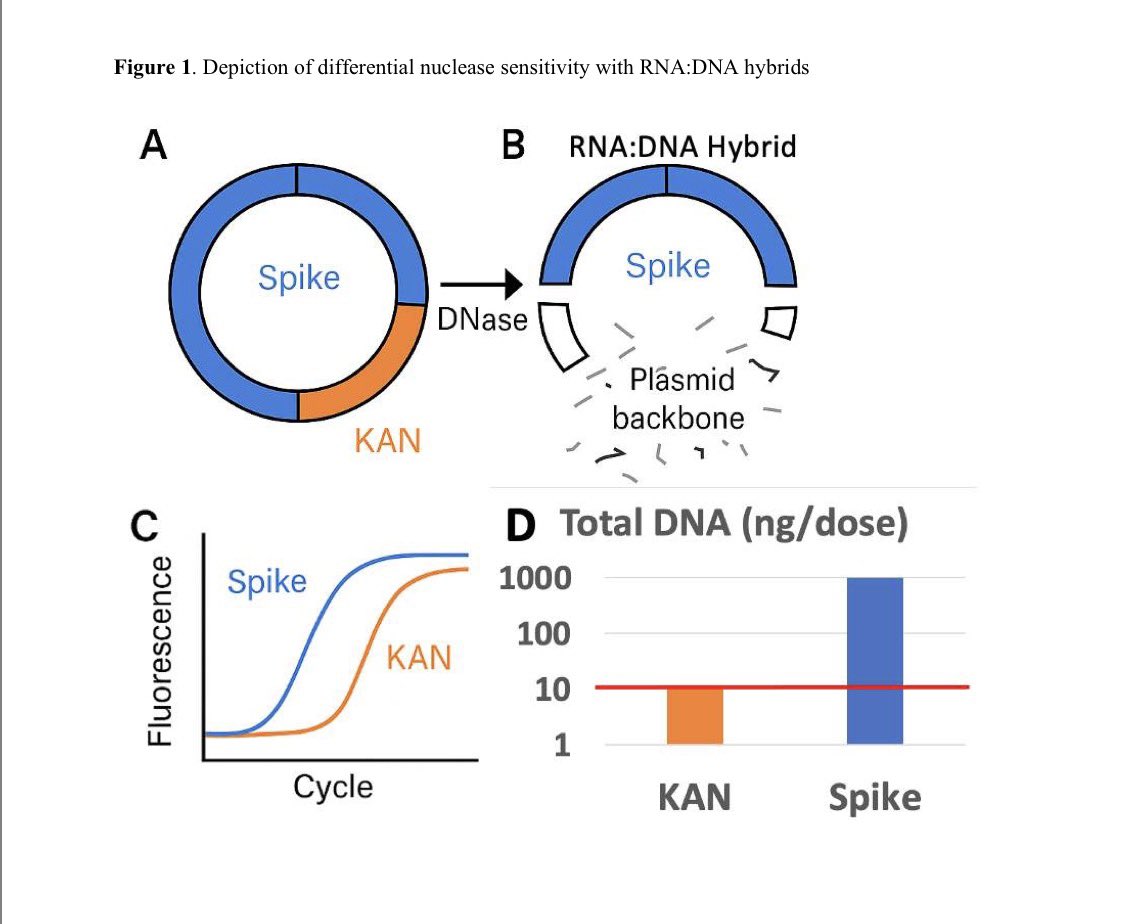
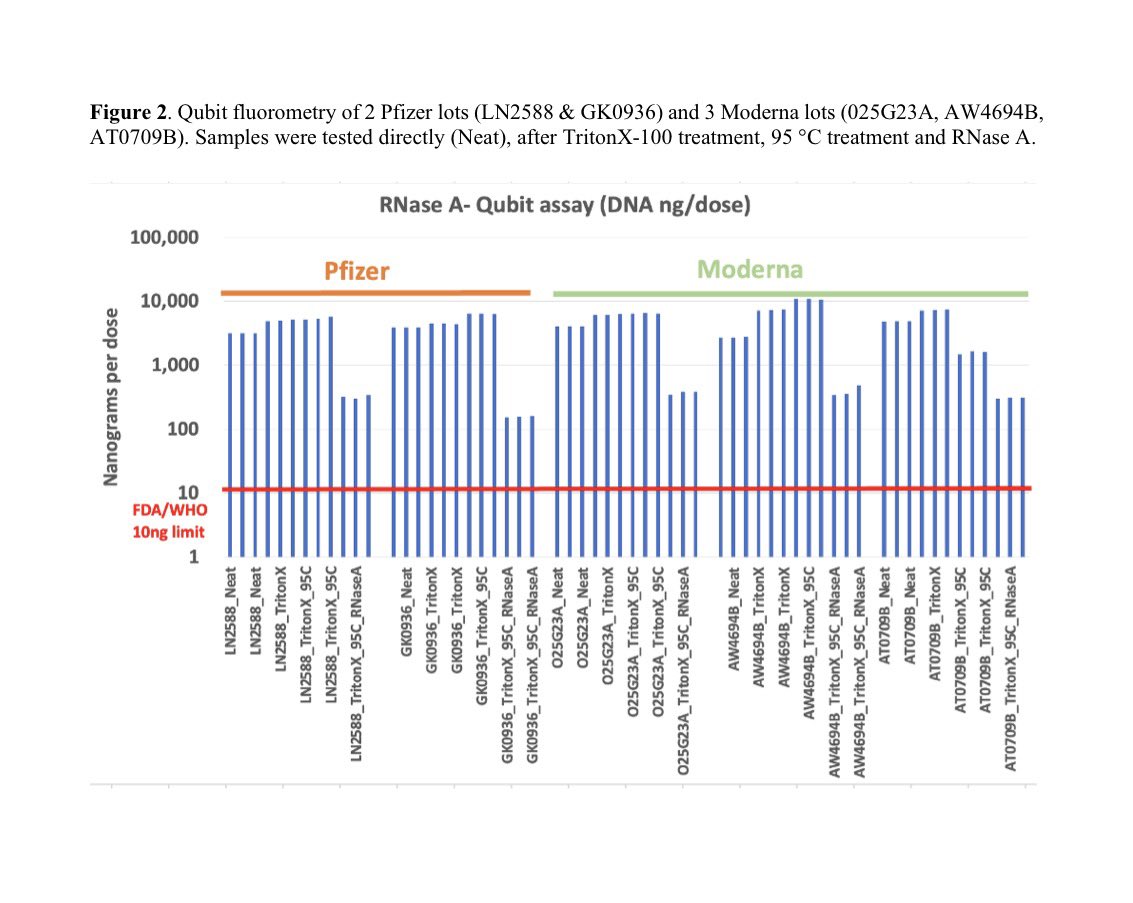
The study was well controlled with RNAse confirmation that the target is in fact DNA and not RT of RNA.
Team "Scientific Censorship" has a lot of explaining to do. Many people on this team were vocal opponents of the CRISPR baby experiments run in China. Now they are guilty of
Team "Scientific Censorship" has a lot of explaining to do. Many people on this team were vocal opponents of the CRISPR baby experiments run in China. Now they are guilty of
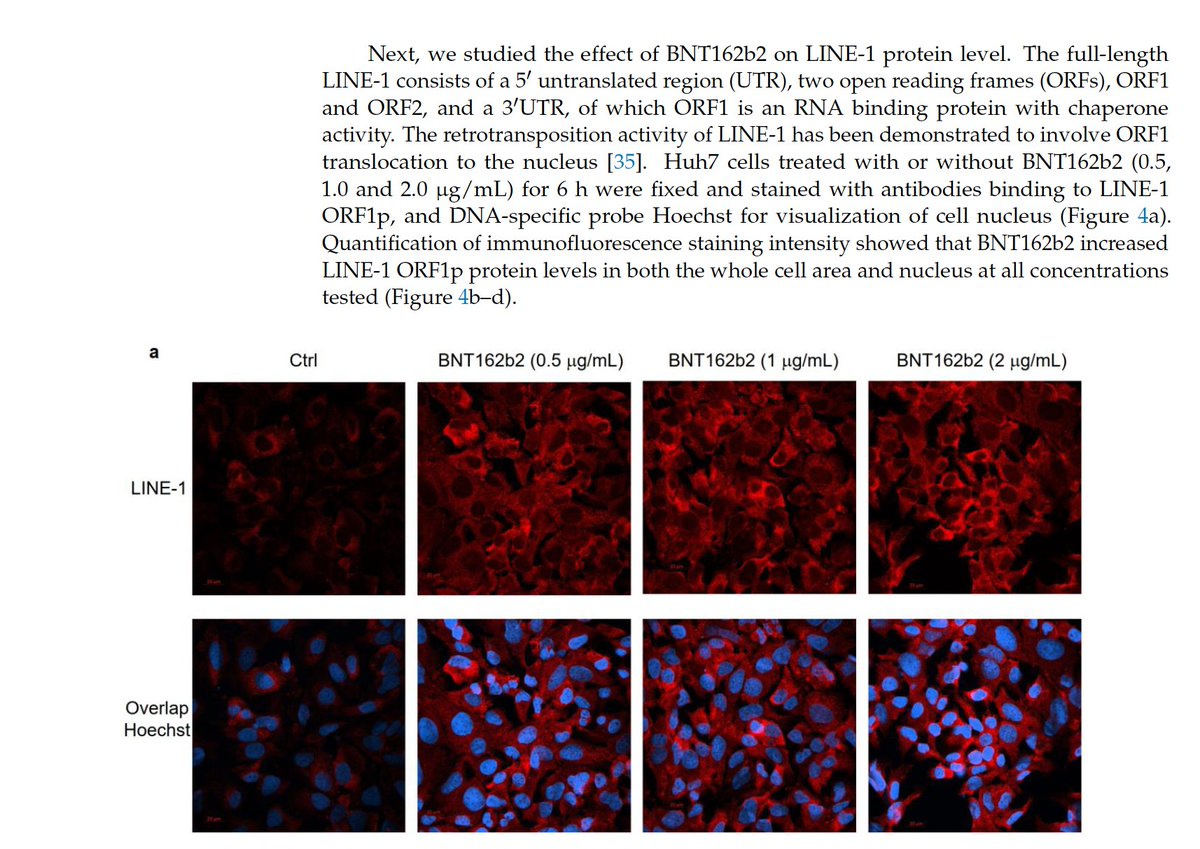
Advocating such experiments on a billion people. It is amazing how pliable their ethics are with a pinch of fear. 
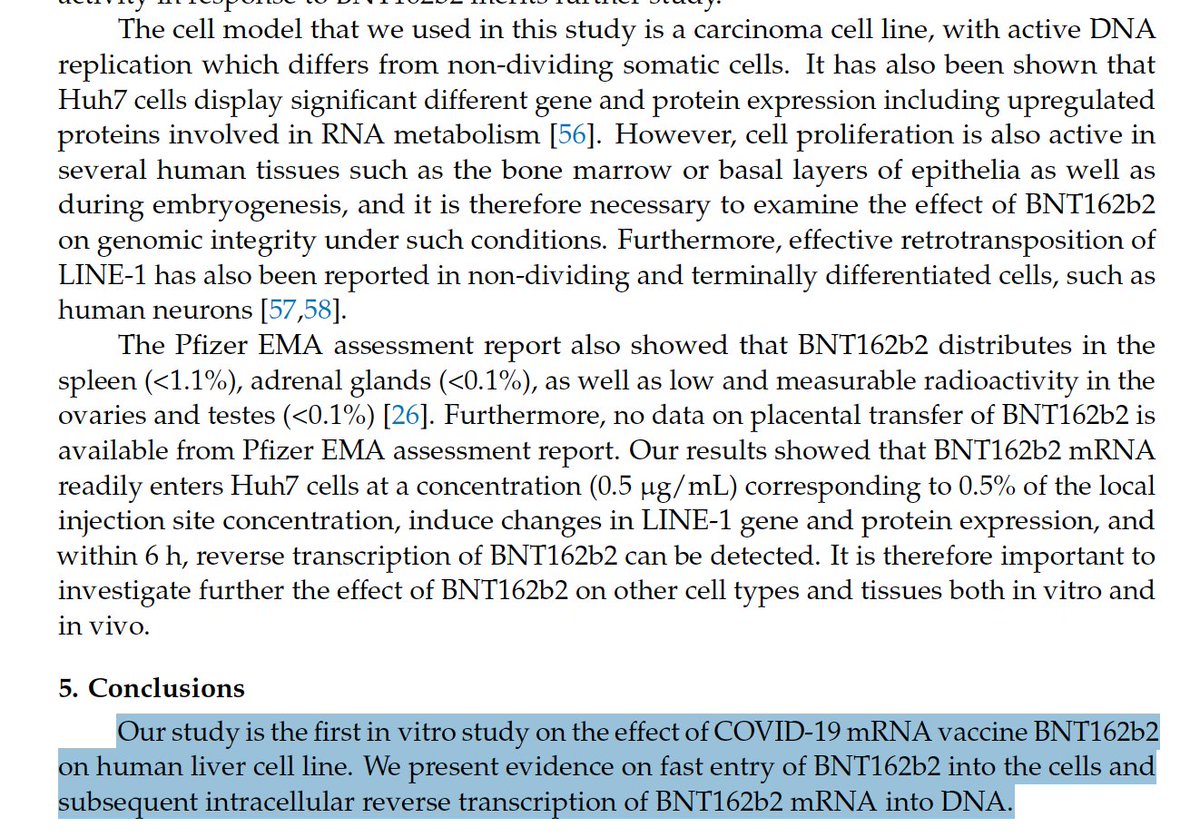
We covered this in our PrePrint with Dr. McCullough.
@P_McCulloughMD
In a sane world this would lead to immediate moratorium on mRNA transfections until WGS are complete.
We are no longer in a sane world and you should take protection into your own hands
osf.io/bcsa6/
@P_McCulloughMD
In a sane world this would lead to immediate moratorium on mRNA transfections until WGS are complete.
We are no longer in a sane world and you should take protection into your own hands
osf.io/bcsa6/
Corrected Link
mdpi.com/1467-3045/44/3…
mdpi.com/1467-3045/44/3…
Adding Rudi's paper to thread so it doesn't get lost in the reply's
https://twitter.com/dimgrr/status/1497303086158778371?s=20&t=YOpnFUNwiYf8K7gmMTT8Ug
Fortunately, Twitter has the receipts of the woke Mob.
Check out the comments on this paper.
biorxiv.org/content/10.110…
Check out the comments on this paper.
biorxiv.org/content/10.110…
A good question from a fellow cat...
When will Whole Genome Sequence Moon?
I think they will need long read sequencing to sort out LINE-1 integration. That usually requires >100ng of DNA.
Human genome is 6pg so 10,000-100,000 cells of DNA are needed to sequence.
When will Whole Genome Sequence Moon?
I think they will need long read sequencing to sort out LINE-1 integration. That usually requires >100ng of DNA.
Human genome is 6pg so 10,000-100,000 cells of DNA are needed to sequence.
Each cell is unlikely to carry integrations in the same location of the genome. So they can't use consensus sequencing (ensemble of all 10,000 cells DNA sequencing) to sort it out. PacBio HiFi sequencing can deliver 150Gb in a single flow cell or 50X coverage per flow cell
More recent studies are pushing this number down to 5ng for PacBio, so they maybe able to get away with 1,000 cells. pacb.com/wp-content/upl…
This might require multiple flow cells. 10 would give you 500X. Either way, this could be over $25K in sequencing but the accuracy of the HiFi reads means each 20Kb read is as good as Sanger sequencing but 30X longer and better for mapping in LINE-1 regions.
My PacBio yield numbers may be dated from the time we ran HiFi on Jamaican Lion.
medicinalgenomics.com/jamaican-lion-…
medicinalgenomics.com/jamaican-lion-…
They improve at a faster than Moore's law rate and they may be able to better speak to current yields. Will need to be done in those Huh7 cell lines and ultimate in patient biopsies.
@PacBio
pacb.com/wp-content/upl…
@PacBio
pacb.com/wp-content/upl…
I did leave a comment in Rudi preprint that suggested DNAse and RNAse controls.
It had some push back.



It had some push back.
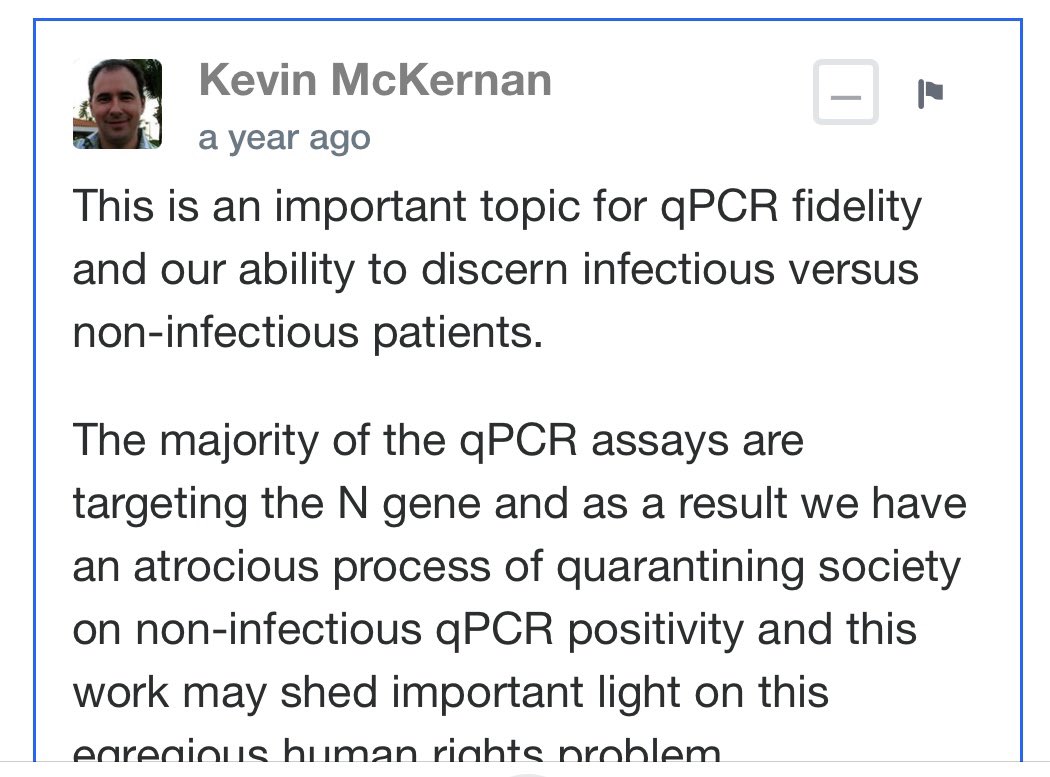



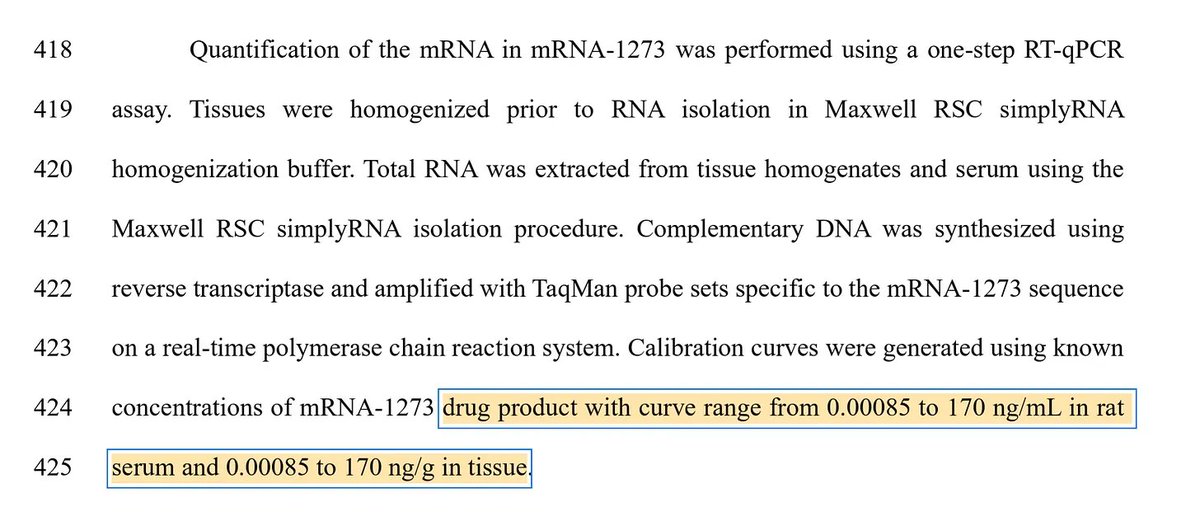
The DNAse and RNAse experiments we just done on BNT162b2 and further move this from conspiracy towards Fact. 

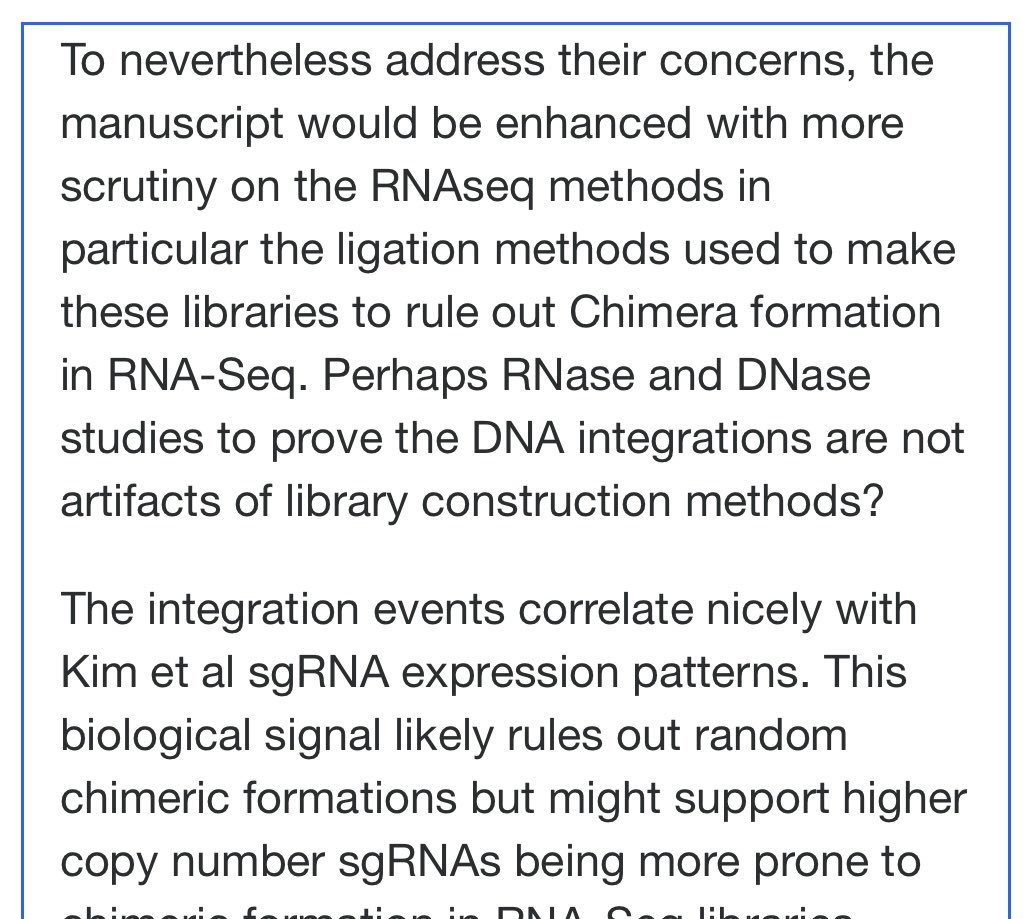
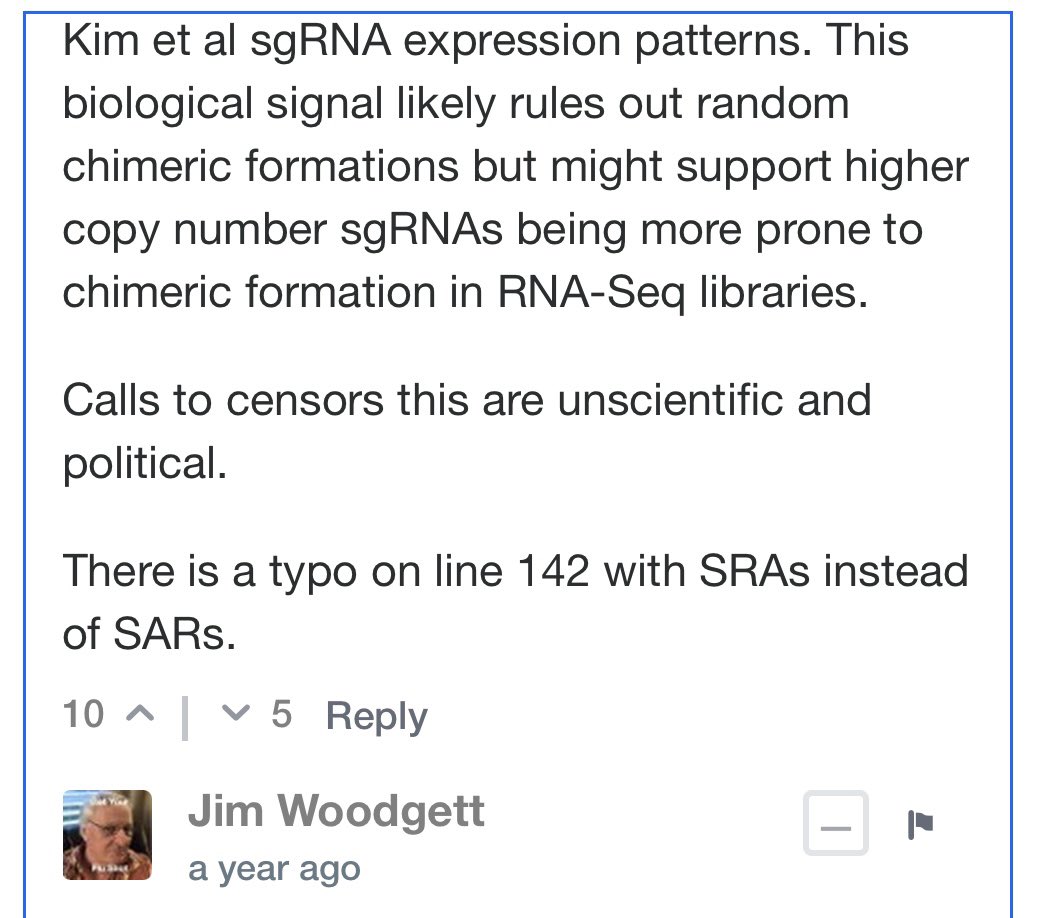
Before we get overly focused on Integration….
We should pause to realize this paper has data that should concern us today.
The paper demonstrates vax induced elevated LINE-1 expression.
Elevated LINE-1 expression is not a good thing.
frontiersin.org/articles/10.33…
We should pause to realize this paper has data that should concern us today.
The paper demonstrates vax induced elevated LINE-1 expression.
Elevated LINE-1 expression is not a good thing.
frontiersin.org/articles/10.33…
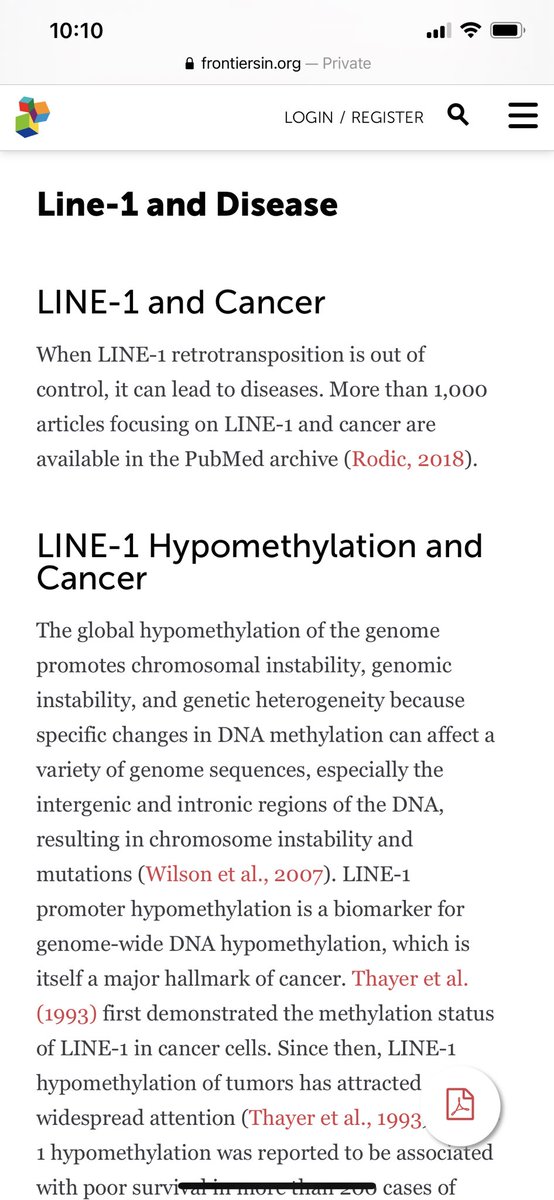
There are a lot of questions regarding if an integration event would even transcribe or just be a genomic fossil.
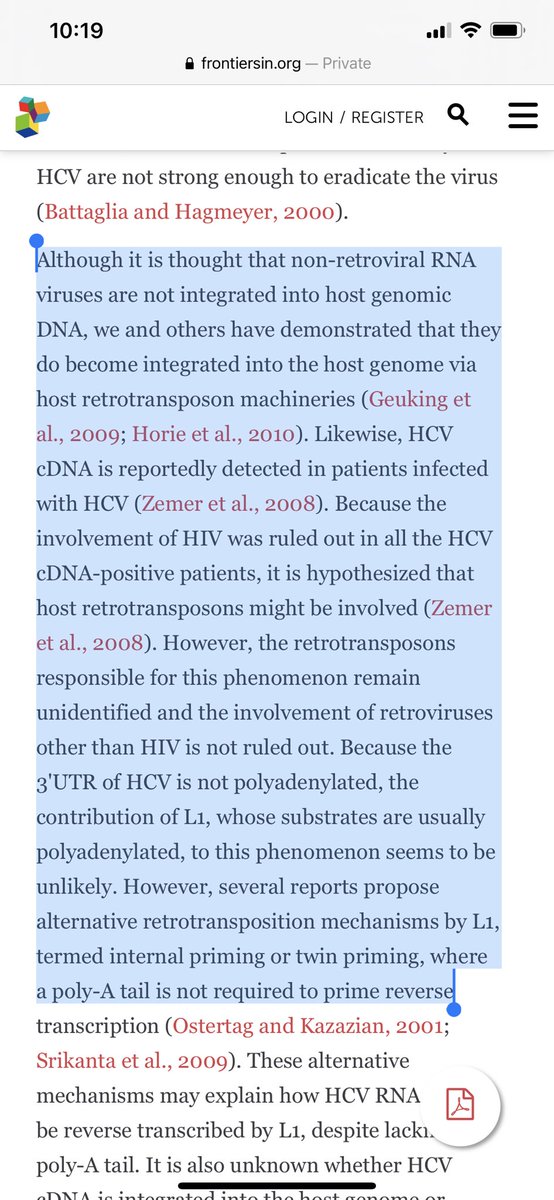
We may learn something from HCV.
frontiersin.org/articles/10.33…
We may learn something from HCV.
frontiersin.org/articles/10.33…
It is worth reading the Back and Forth on the Jaenisch labs paper (Zhang et al).
They suggest integration with the virus is 2-5 integrations per 10,000 cells.
pnas.org/doi/full/10.10…
They suggest integration with the virus is 2-5 integrations per 10,000 cells.
pnas.org/doi/full/10.10…
@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh