With three Omicron-related recombinants getting assigned Pango lineages I'm going to write a thread covering:
- What are recombinants?
- Why are we seeing so many now?
- What exactly are the new lineages XD, XE and XF?
- How concerned should we be about them?
- What are recombinants?
- Why are we seeing so many now?
- What exactly are the new lineages XD, XE and XF?
- How concerned should we be about them?
First off, what are recombinants?
When two related viruses infect the same cell (ie during a coinfection) the viral replication machinery can accidentally switch from one genome to the other resulting in a mixed genome - this is viral recombination.
When two related viruses infect the same cell (ie during a coinfection) the viral replication machinery can accidentally switch from one genome to the other resulting in a mixed genome - this is viral recombination.

SARS2 has been doing this all of the way through the pandemic - however its only easy to see when the two parental viruses are distantly related (for example Alpha/prevariant in the paper below)
sciencedirect.com/science/articl…
sciencedirect.com/science/articl…
Therefore the reason we are seeing lots of recombinants is, until recently, we had lots of genetically distinct viruses circulating - Delta, BA.1 and BA.2 - when recombinants occur between these lineages it is possible to identify, even if the switched part of the genome is small
The other reason we are seeing a lot now is because when BA.1 took off (at least in Europe and the USA) there was very high levels of Delta already circulating (see UK below) - therefore lots of opportunities to co-infect, recombine and transmit onwards. 

So what are the new recombinant lineages? They fall into 2 catagories:
- Delta x BA.1 (unhelpfully refered to as Deltracron in the media... journalists please stop doing this!)
- BA.1 x BA.2
- Delta x BA.1 (unhelpfully refered to as Deltracron in the media... journalists please stop doing this!)
- BA.1 x BA.2
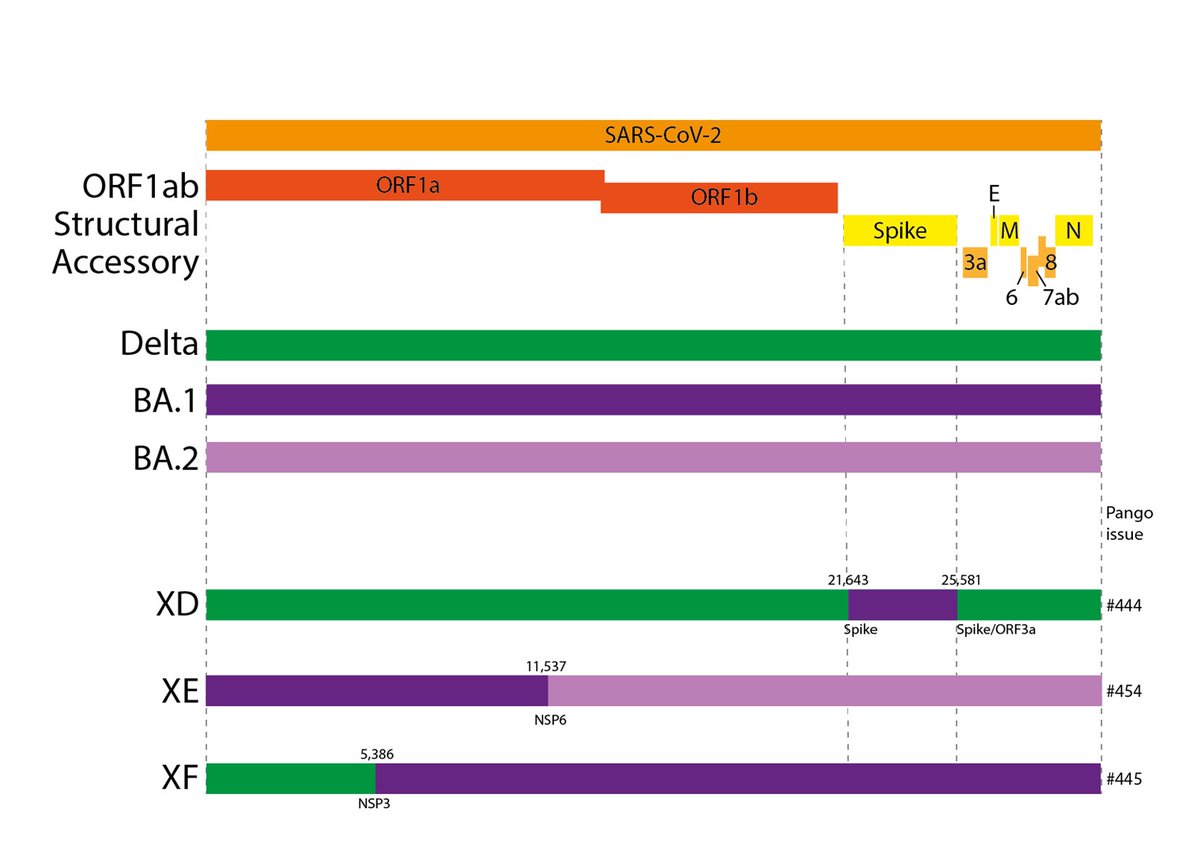
XD is the new name for the French Delta x BA.1 lineage. Sequenced and verified by @SimonLoriereLab and colleagues at Pasteur, first spotted by @DrSnOU. It contains the Spike protein of BA.1 and the rest of the genome from Delta. It currently comprises several 10s of sequences. 

XE is a large UK BA.1 x BA.2 lineage. Sequenced and verified by @UKHSA and @sangerinstitute, first identified by @nzm8qs. It has the Spike and structural proteins from BA.2 but the 5' part of its genome from BA.1. It comprises several hundred sequences at present. 

XF is a UK Delta x BA.1 lineage. Again sequenced and verified by @UKHSA and first identified by @nzm8qs. It has the Spike and structural proteins from BA.1 but the 5' part of its genome from Delta. It comprises several tens of sequences currently. 

(Just for a comparison, the UK currently uploads ~~10x more sequences than France therefore 10s of sequences in France are roughly equivalent to ~~100s of sequences from the UK)
So how concerned should we be about these (and other) recombinants?
Recombinants that contain the Spike and structural proteins from a single virus (like XE or XF) are fairly likely to act similarly to thier parental virus - nearly all we understand about variants maps here.
Recombinants that contain the Spike and structural proteins from a single virus (like XE or XF) are fairly likely to act similarly to thier parental virus - nearly all we understand about variants maps here.

XD is maybe a little more concerning... It has been found in Germany, Netherlands and Denmark and it contains the structural proteins from Delta - if *any* of these recombinants were to act much differently than its parent it might be XD... 

All these recombinants (including the smaller clusters that have not been assigned) should clearly be closely monitored for signs of growth and attempts should be made to isolate and characterise where possible...
Below is a full list of the currently verified recombinants
Below is a full list of the currently verified recombinants

Also congratulations to @SimonLoriereLab and colleagues at Pasteur, and scientists at IHU who have both managed to independently isolate the XD recombinant - will be very interesting to see how the virus behaves compared to its parental lineages.
Also recommend this nice thread by @alexbolze describing some recombinants and co-infections they found during surveillence in the USA
https://twitter.com/alexbolze/status/1502861663199903748
Finally, special mention to the @GISAID contributors and the @PangoNetwork community, particularly @nzm8qs, who is a one man recombinant-spotting machine!
Links to the pango github page with more details on XD, XE and XF:
github.com/cov-lineages/p…
github.com/cov-lineages/p…
github.com/cov-lineages/p…
github.com/cov-lineages/p…
github.com/cov-lineages/p…
github.com/cov-lineages/p…
(also just to say it will take a short while for these lineages to get updated on sites like outbreak. info, gisaid, covspectrum, etc)
Update 1: Preprint from US CDC on discovery and verification of the USA Delta x BA.1 recombinant (see below)

https://twitter.com/biorxivpreprint/status/1506081450344566789

Update 2: several further BA.1 x BA.2 recombinants have been assigned by Pango. Interestingly all have breakpoints in ORF1ab.
- XG is the largest (Mostly in Denmark), ~120 sequences
- XH, XJ, XK, XL are all currently <100 sequences
- XG is the largest (Mostly in Denmark), ~120 sequences
- XH, XJ, XK, XL are all currently <100 sequences

...and from UKHSA tech briefing (as @kallmemeg describes below) XE currently appears to be showing a modest growth advantage over BA.2 in the UK (interesting to see if this holds true!)
https://twitter.com/kallmemeg/status/1507394818871336960
Small warning here - suspect the first 3 omicron X lineages will start feeding onto the major platforms/databases this week (outbreak, gisaid, covspectrum, etc) - its quite likely there will be a few teething issues with seqs getting misassigned, etc, best to double check first!
Also for recombinant hunters - very highly recommend this great thread by @nzm8qs - who has probably spotted more recombinant lineages than almost everyone else put together! - on his methods for finding recombinants!
https://twitter.com/nzm8qs/status/1509225414589255683
Small Friday update - couple more European BA.1 x BA.2 recombinants designated - XM and XN:
XM - mainly Netherlands and Germany, ~60 sequences currently (spotted by @JosetteSchoenma)
XN - another UK recombinant, ~100 sequences currently (spotted by @nzm8qs)
XM - mainly Netherlands and Germany, ~60 sequences currently (spotted by @JosetteSchoenma)
XN - another UK recombinant, ~100 sequences currently (spotted by @nzm8qs)

Another few recombinant lineages got assigned yesterday as well. Here is the current list of assigned BA.1 x BA.2 recombinants. Includes 2 more UK recombinants (XP, XQ & XR - all spotted by @nzm8qs). XP is interesting as it actually has the Spike from BA.1.1 (ie R346K) 

And here is the current list of notable Delta x BA.1 recombinants. XS is the new addition from the USA (spotted by @nzm8qs). It does rather look like these are mostly getting outcompeted by BA.2 as far as we can tell.... 

No new X lineages this week (yet) but a couple of minor updates.
UKHSA variant tech briefing showing a continued growth rate advantage of XE over BA.2* of between 10-20% (similar to last years AY.4.2 over Delta). Data is still fairly preliminary though.
gov.uk/government/pub…
UKHSA variant tech briefing showing a continued growth rate advantage of XE over BA.2* of between 10-20% (similar to last years AY.4.2 over Delta). Data is still fairly preliminary though.
gov.uk/government/pub…

..and a brilliant preprint from @SimonLoriereLab and colleagues at the Pasteur characterising the Delta x BA.1 recombinant XD. Strikingly XD shows an intermediate pathogenicity phenotype between BA.1 and Delta in K18 mice:
researchsquare.com/article/rs-150…
researchsquare.com/article/rs-150…

This shows some of the lower severity seen in the mouse model of BA.1 maps *outside* the Spike protein which is really interesting.
Fortunatley (for us) XD appears to mostly have been outcompeted by BA.2, but a similar recombinant arising with a BA.2 spike is a worry.
Fortunatley (for us) XD appears to mostly have been outcompeted by BA.2, but a similar recombinant arising with a BA.2 spike is a worry.
I let this thread fizzle out before as most of the recombinants were getting outcompeted by BA.5, but briefly reviving to to highlight a couple of pretty interesting 'complex' recombinants that recently got assigned - XAY and XAW. Both of which are BA.2 x Delta recombinants. 

XAY (and its sister recombinant - the minor, currently unassigned 'constellation 2') have both been found in South Africa - they are unique in that they are clearly related but have a very complex pattern of recombination compared to previous recombinants
https://twitter.com/Tuliodna/status/1563192408988196864
XAW is slightly different - its a fairly standard 'sandwich' recombinant (Delta bread and BA.2 filling), reminiscent of XD with the spike from BA.2 and most of the rest of the genome from Delta. 

Both XAY and XAW have a number of additional RBD mutations on top of BA.2 that probably impact antigenicity - however both are currently at very low seq numbers currently (<10). Most likely they end up as oddities but lots of eyes are wathcing closely to see if that changes...
The number of additional mutations combined with the usual parent lineages (i.e. Delta and BA.2 which didnt ever co-circulate much in most places) leads us to hypothesis these might have arisen from chronic co-infections (chronic Delta infection, co-infected with BA.2?)
• • •
Missing some Tweet in this thread? You can try to
force a refresh