
Virologist. Influenza and Coronaviruses. Mostly not on X. Find me on the other site.
20 subscribers
How to get URL link on X (Twitter) App


https://twitter.com/PeacockFlu/status/1682696057744916480What I'm not really able to cover yet is the North American cattle situation - not enough sequencing or epidemiological data has been shared to draw any strong conclusions - see this recent piece by @HelenBranswell This is frustrating to say the least...
https://twitter.com/statnews/status/1781365515299996050

 Swine influenza viruses have recieved a bit of attention recently - with 'cryptic' (ie no know contact with pigs) infections found in the UK and the Netherlands in the last few months
Swine influenza viruses have recieved a bit of attention recently - with 'cryptic' (ie no know contact with pigs) infections found in the UK and the Netherlands in the last few months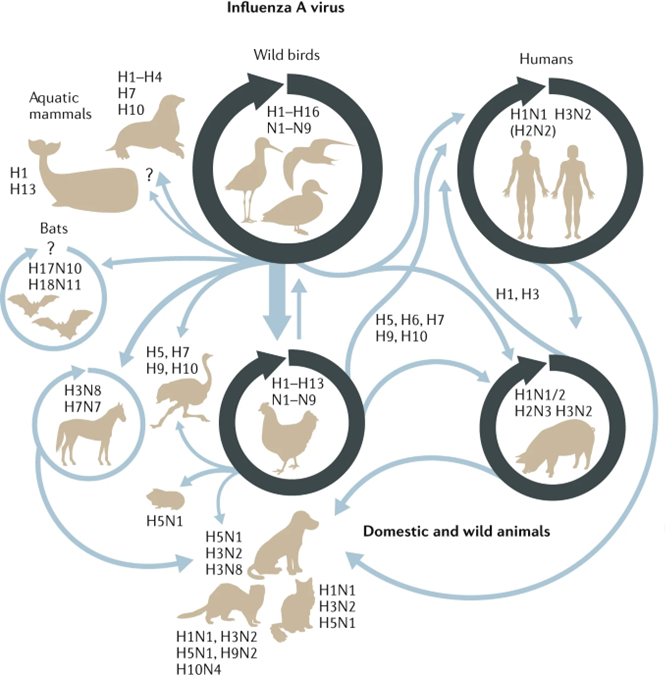
 Firstly, a quick situational update on the panzootic in birds. We're now 3 years into this outbreak and the virus is continuing to spread across the world, largely impacting waterfowl and seabirds (including many that are endangered)
Firstly, a quick situational update on the panzootic in birds. We're now 3 years into this outbreak and the virus is continuing to spread across the world, largely impacting waterfowl and seabirds (including many that are endangered) https://twitter.com/guardianeco/status/1681980535843999744

https://twitter.com/NaturePortfolio/status/1674077465885519874First off we did find some sarbecoviruses (distantly related to sars1 and 2) that had detecatable human ace2 binding, however this was pretty weak. We also know that it doesnt take that much go switch from weak to strong binding with sarbecos though.


https://twitter.com/virological_org/status/1651202114062876678What does ORF8 do?




https://twitter.com/BenjMurrell/status/1570862197827567620
https://twitter.com/yunlong_cao/status/1577343557048098818


https://twitter.com/ImperialInfect/status/1538808398187319296Some context - the last few years have seen mass wild bird die offs and some of the highest numbers of jumps into poultry farms ever seen across Europe (in particular in the UK) nature.com/articles/d4158…



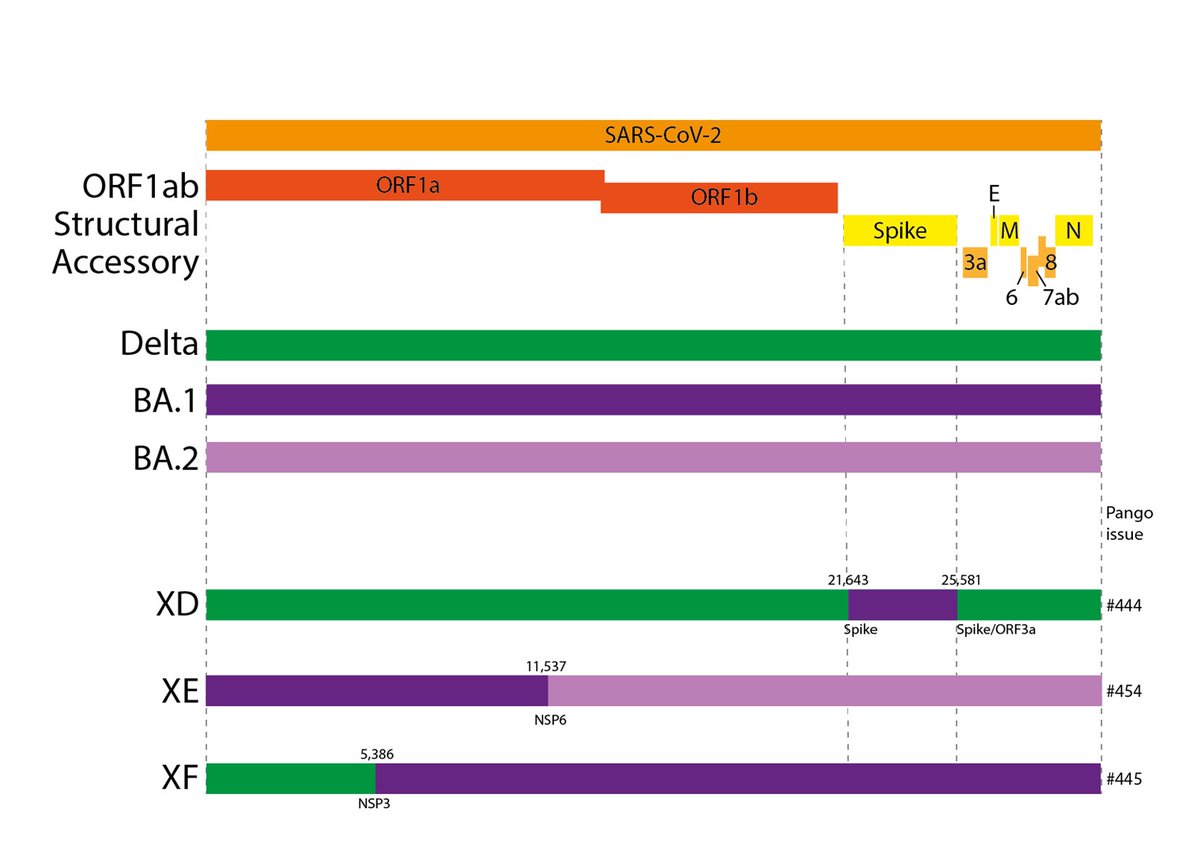
 First off, what are recombinants?
First off, what are recombinants?

https://twitter.com/BioWilko/status/1499359600113553410One of the mutations that repeatedly, independently, arises during known chronic infections is E:T30I, after Spike: E484K, it is the most common mutation we saw in this dataset. (though worth saying it arises during *many*, but not *all* chronic infections)


https://twitter.com/CellReports/status/1491135252525314048Here's a thread I made when it preprinted summarising the paper for those interested in the full details
https://twitter.com/PeacockFlu/status/1429005642254139393
https://twitter.com/shay_fleishon/status/1480934070012059649

https://twitter.com/shay_fleishon/status/1480086456022577153In comparison the actually real recombinant lineage XC (an Alpha/Delta recombinant found in Japan a few months back) forms a really nice, single cluster (in red)
