We're now starting to see the evolution of new potentially impactful sublineages of Omicron with particular focus on mutations at spike residue 452. Here, I'd like to highlight lineages B.2.12.1 in New York, as well as BA.4 and BA.5 in South Africa. 1/17
Stepping back, Omicron emerged as three distinct lineages BA.1, BA.2 and BA.3 and despite the head start of BA.1, we've seen BA.2 overtake BA.1 across the world over the course of January to April. 2/17 

Variant "fitness" will depend on intrinsic transmissibility and escape from existing population immunity. The first tranche of variants (Alpha, Delta, etc...) largely spread due to increased intrinsic transmissibility, while Omicron spread primarily due to immune escape. 3/17
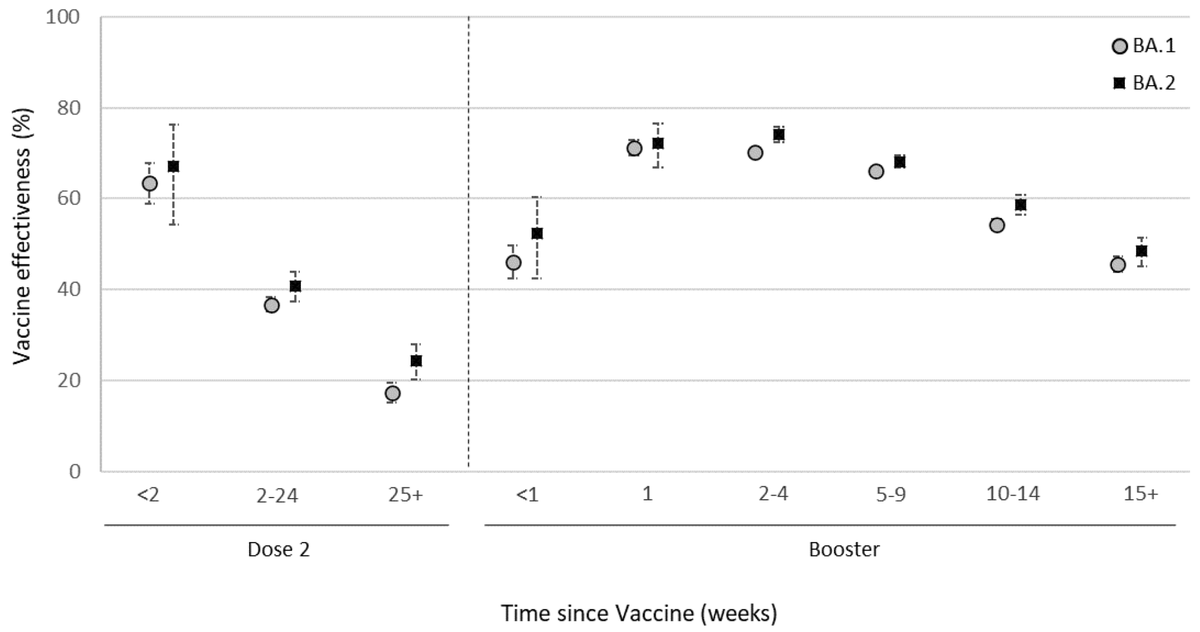
BA.2's advantage over BA.1 appears to be due to intrinsic transmissibility. The antigenically important S1 region of spike is highly similar between BA.1 and BA.2 and we observe similar vaccine effectiveness between BA.1 and BA.2 (figure from @UKHSA gov.uk/government/pub…). 4/17 
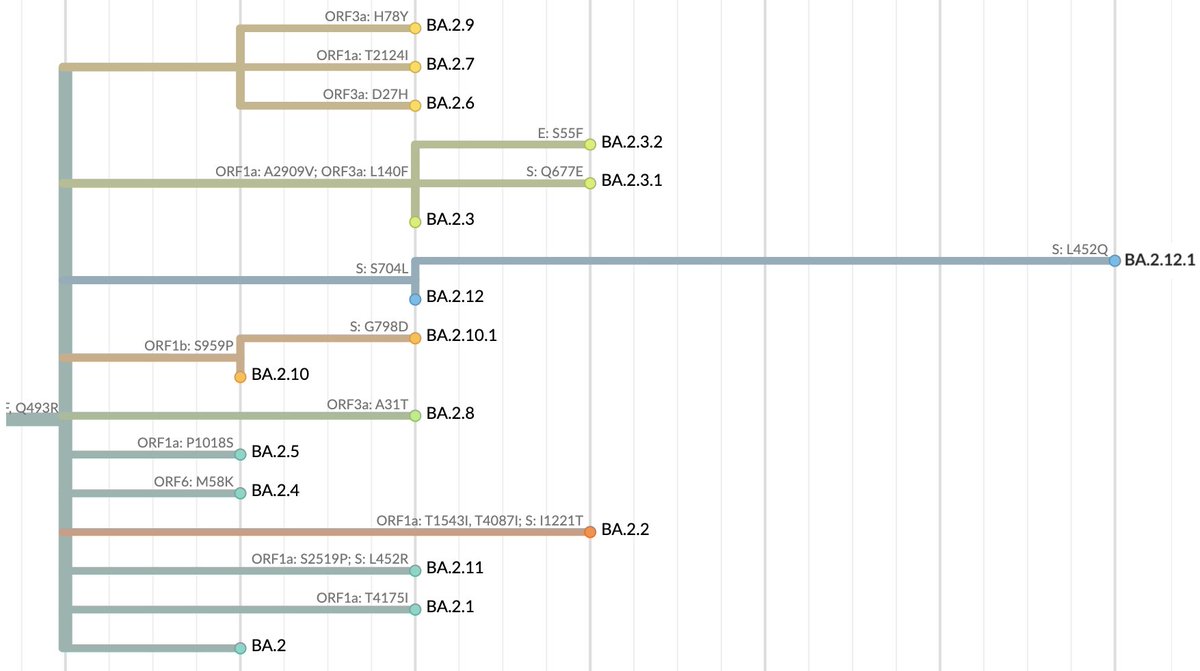
With BA.2's global dominance we expect further evolution to occur on top of BA.2. @PangoNetwork now classifies 21 sublineages of BA.2. However, most of these sublineages are characterized by mutations thought to have little impact (figure from nextstrain.org/nextclade/sars…). 5/17 
Here, the one to watch just based on mutations is B.2.12.1 which has spike mutations S704L and L452Q on top of BA.2 background. Previously, L452R appeared to have an important role in promoting the spread of Delta and also showed up in Epsilon and Lambda. 6/17
The primary geographic focus for B.2.12.1 is New York state, which was up to ~18% frequency as of April 1 from ~1% at the beginning of March. Similarly, Massachusetts has increased to 7% frequency as of April 1 from below 1% frequency at the beginning of March. 7/17 
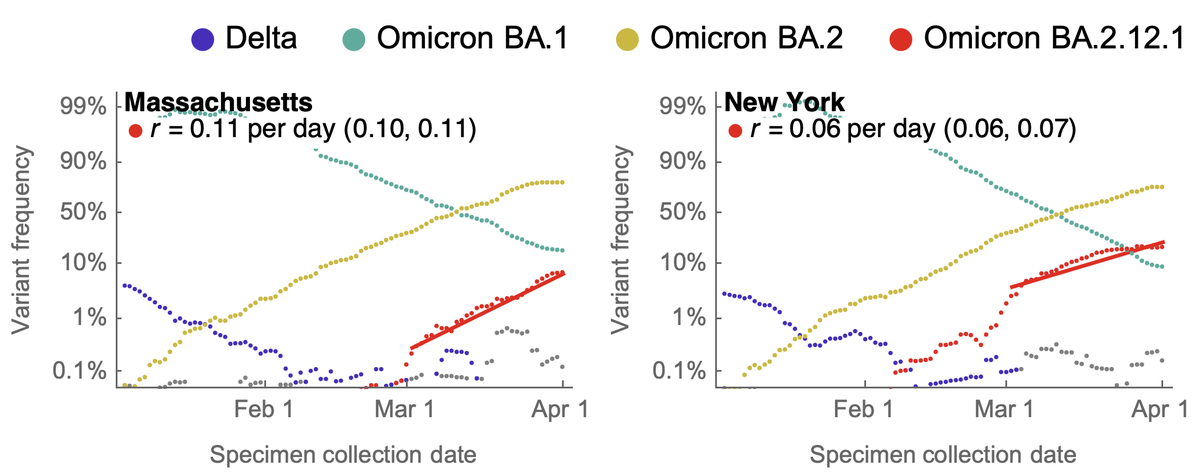
We observe a logistic growth rate of 0.06 per day in NY and 0.11 per day in MA. This is similar in magnitude to the observed advantage of BA.2 over BA.1. 8/17
We have a similar, though somewhat more complex, situation in South Africa with lineages BA.4 and BA.5 which share spike mutations L452R, F486V and reversion Q493R. Figure from @CorneliusRoemer 
https://twitter.com/CorneliusRoemer/status/1514255166584893446. 9/17
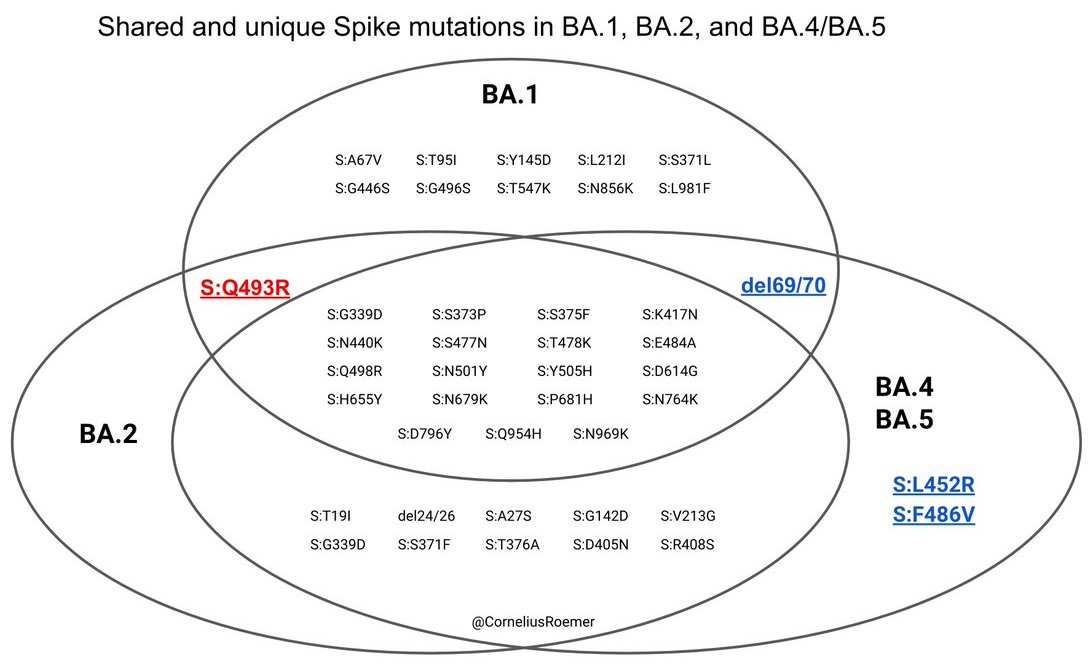
It's not entirely clear to me whether BA.4 and BA.5 are sister lineages to BA.2 or sublineages of BA.2, but this distinction shouldn't matter for assessment of impact on viral circulation. 10/17
The primary geographic focus for BA.4 is Gauteng, where as of April 1 it was at ~60% frequency, and the primary focus for BA.5 is KwaZulu-Natal, where as of April 1 it was at ~55% frequency. 11/17 
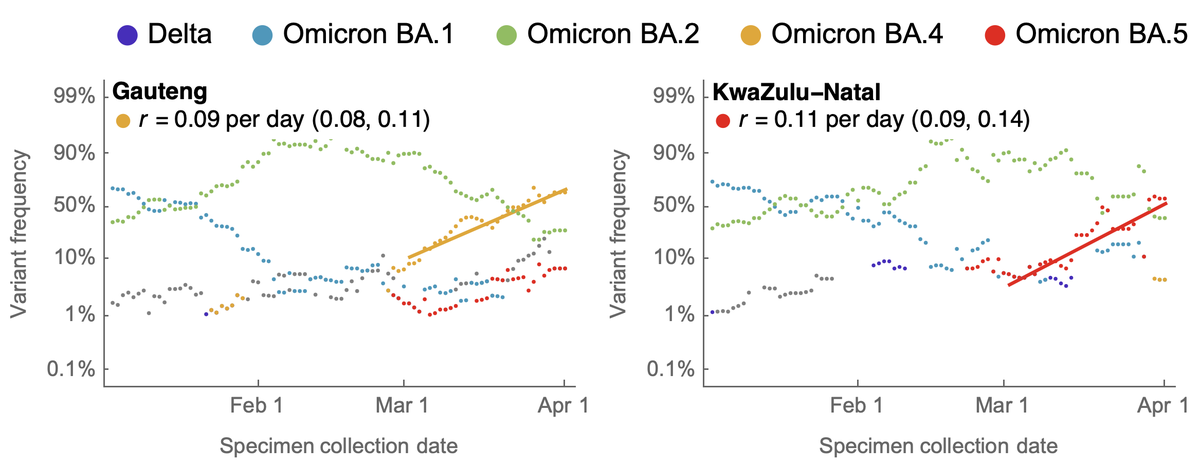
We observe logistic growth rates of BA.4 in Gauteng of 0.09 per day and of BA.5 in KwaZulu-Natal of 0.11 per day. Again, this is similar in magnitude to the observed advantage of BA.2 over BA.1. 12/17
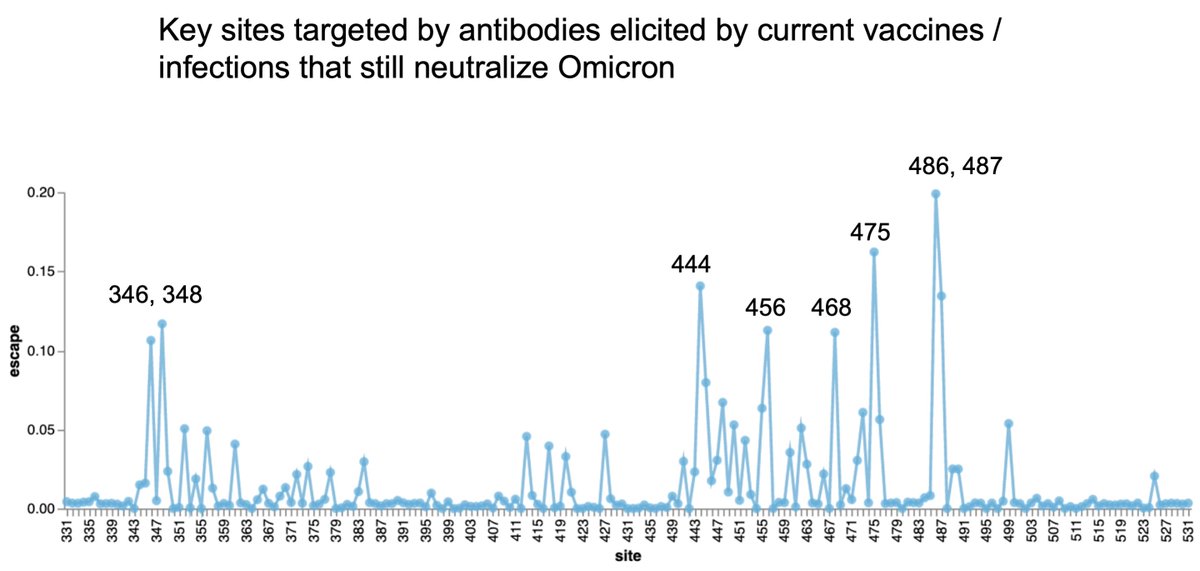
Based on serological experiments it seems quite likely that 486V would confer some additional immune escape relative to BA.2 (figure from @jbloom_lab 
https://twitter.com/jbloom_lab/status/1513538000432472069). However, 452R/Q is not obviously a key antigenic mutation. 13/17
The hypothesis is then that 452R/Q is conferring some additional intrinsic transmission advantage. Distinguishing these scenarios is challenging however and largely relies on assessing neutralization titer in assays with 452R/Q viruses and recent human sera. 14/17
Looking forward, I expect these 452R/Q sublineages to continue to expand. As they do so, they may acquire additional mutations with the "winning" sublineage the one that accumulates the best constellation of mutations. 15/17
However, it's also possible for some other mutational constellation to arise (on top of BA.2 or otherwise) and overtake these new 452R/Q lineages. 16/17
https://twitter.com/trvrb/status/1511871614521995269
This sort of accumulation of mutations that drive further host adaptation and antigenic drift is my general expectation for evolution in the coming months. It's possible we may have additional "Omicron-like" events, but my baseline is this steady "flu-like" scenario. 17/17
Follow up #1: And of course, I made a consistent typo in the thread where I write "B.2.12.1" when instead I should have written "BA.2.12.1". The figures are correctly annotated at least.
Follow up #2: The above was remiss in not acknowledging the incredible work of @Tuliodna, @ceri_news, @nicd_sa, @nhls_sa in generating and sharing sequence data on BA.4 and BA.5 in South Africa. They've been absolutely essential throughout the pandemic.
https://twitter.com/Tuliodna/status/1513529930046648326
• • •
Missing some Tweet in this thread? You can try to
force a refresh