
Excited to share that our @NatureBiotech paper with Aviv Regev on Multicellular Programs (MCPs) is now out with a fully automated data-driven method to identify MCPs from #singlecell or spatial data #behindthepaper: go.nature.com/3iQ8fgW nature.com/articles/s4158… (1/19) 

Gene programs are often studied within a cell, with each cell considered an “independent entity”. However, cells from the same niche/tissue/organism are not independent, but share external signals, genetic background/lineage, and can directly impact one another. (2/19) 

Here we define “Multicellular Programs (MCPs)” as different sets of genes working in concert across different cell types. (3/19) 
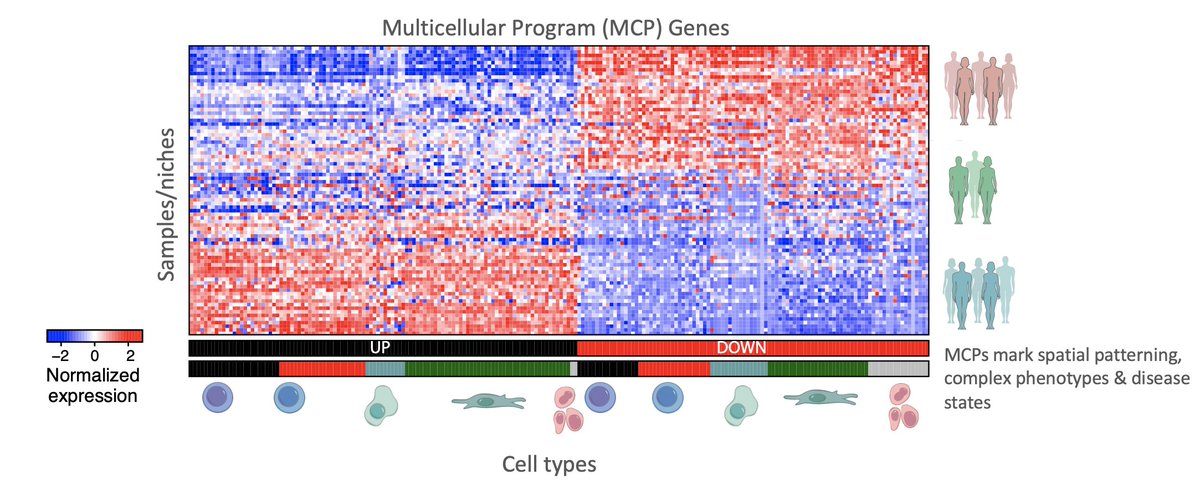
Then we developed DIALOGUE – a dimensionality reduction method to robustly identify MCPs from single cell/spatial data. (4/19)
How does DIALOGUE work? Given #singlecell or #SpatialTranscriptomics data, DIALOGUE treats different cell types from the same micro/macroenvironment or sample as different representations of the same entity and identifies MCPs in two steps. (5/19) 

Step I. DIALOGUE identifies sparse canonical variates that transform the original feature space to a new feature space, where the different, cell-type-specific, representations are correlated across the different samples/environments. (6/19)
Step II. DIALOGUE then fits multilevel mixed-effects models to identify the specific genes that comprise these latent features, accounting for single-cell distributions and confounders. (7/19)
The key is that DIALOGUE needs to look at cells that are in the same niche. When we have spatial data, that can be direct proximity or the same area. And when we have single cell data, this can be cells that are in the same sample. Here’s how it performed in each case. (8/19)
DIALOGUE shows that MCPs dominate spatial patterning, allowing us to predict the expression of one cell from the expression of its neighbors, predict cell location based on the cell’s own transcriptome and spot cells that are “newcomers” or “unrelated” to the environment. (9/19)
DIALOGUE outperformed other methods and spatial predictors, including neural network and hidden Markov random field-based approaches. (10/19)
Applied to MERFISH data from mouse hypothalamus DIALOGUE identified multicellular programs that mark animal behavior and prior learning experiences. (11/19) 

Applied to lung cancer Spatial Molecular Imager data (from @nanostringtech), DIALOGUE identified a program spanning T cells, fibroblasts, and macrophages, recapitulating coordinated response to IL1 and JAK/STAT signaling at the contact points with malignant cells. (12/19) 

MCPs also dominate variation in #scRNA-Seq data and can be found based on co-variation across samples. We show that such MCPs are tightly linked to complex phenotypes including disease states, genetic risk factors, aging, response to immunotherapies and more! See below. (13/19)
DIALOGUE identified an ulcerative colitis multicellular program that includes multiple IBD risk genes previously identified by #GWAS. The program predicted both disease status and treatment response in an independent cohort. (14/19) 

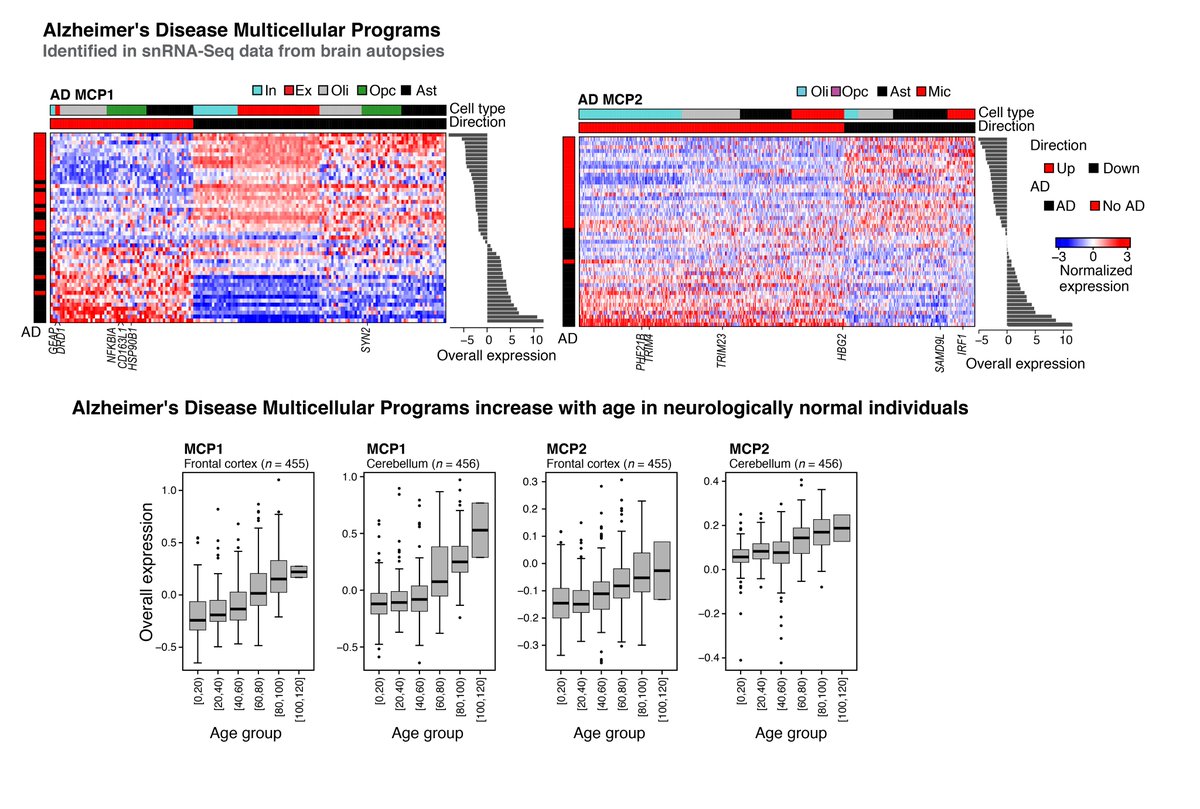
DIALOGUE identified multicellular programs marking #Alzheimers’s disease in #ROSMAP data, which also marked (non-linear) brain aging in a separate cohort from hundreds of neurologically normal subjects. (15/19) 
DIALOGUE identified a multicellular #immunotherapy resistance program in melanoma tumors, linking T cell dysfunction with M2 polarization and APOE repression in macrophage. (16/19) 

The melanoma program provides an additional perspective to the connection between germline APOE genetic variation and immunotherapy resistance and additional support to clinical trials currently combining LXR/ApoE activators with #immunotherapy. (17/19)
These findings demonstrate the power of this approach and perspective in diverse biological settings, study designs, and data types. Check out DIALOGUE and see if you can find MCPs in your data. github.com/livnatje/DIALO… (18/19)
What's next? Our approach can be used to study the nexus between cellular and tissue biology from existing data, and, in the near future, be combined with CRISPR/perturbation screens to identify tissue remodelers and new ways to engineer multicellular circuits! (19/19)
• • •
Missing some Tweet in this thread? You can try to
force a refresh



