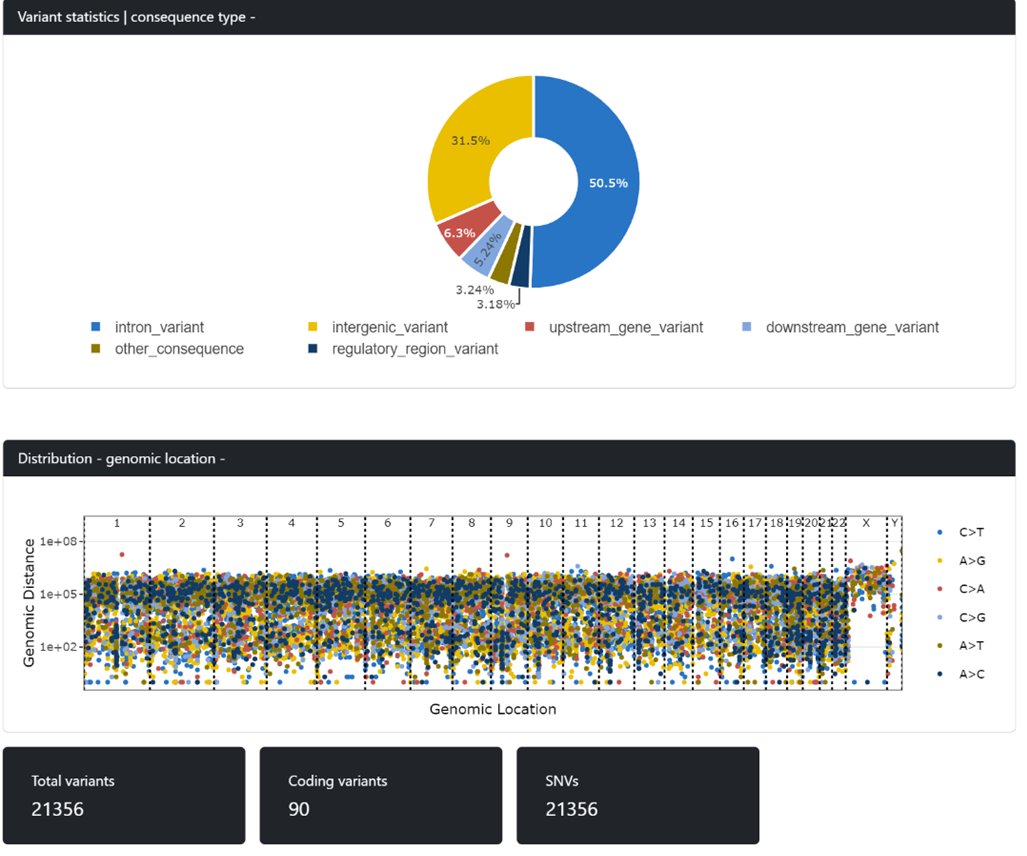
So, as promised, here are the observations of @beggs_lab at @unibirmingham about the new LSK114 kit on the R10.4.1 "Marathon" flow cells - we're happy enough that we have put 4 samples on this afternoon on our @nanopore P24 - ready for London Calling 22 - wet lab by @JoStockton1 

Firstly - output - this is just a representative example but on our tumour samples we are seeing between 80-120gbases of data generated per sample. This is with LSK114 prep, Covaris sheared to ~10kb. Run for 72hrs. Note the data output - much higher thanks to Marathon flow cells 

As you can see we have got roughly the read length we want, and we have found as you increase the read length, data output drops (obvs). Longest read in this run was 668kb. 

Now onto some fun stuff... here is the Fragile X FMR1 gene - v. diff to sequence and top panel shows short reads which completely fail to resolve it (125PE). Below is the LSK114 kit which not only resolves it but detects the 75 base triplet repeat expansion (shown by the purple) 

Just for context, short read really struggles here, but this has seen it, straight off, with no post-processing, and measured the repeat expansion to the single base length - it's a normal length by the way @fragilexuk
Here's another favourite - the BMP7 gene involved in colorectal crypt formation (purple I below) - there's a 213bp *inframe* insertion detection - and it's genuine - we just wouldn't see this in SR seq - and it is going to critically important in cancer formation in this sample 

What's *really* interesting is that the SR data and alignment misinterprets this inframe insertion as a heterozygous SNP - how much more are we missing with short reads?
Another favourite of mine - HLA-C - notoriously difficult to sequence and the short read shows lots of errors, mainly because of alignment difficulties - but the long reads here work perfectly and give great data *in single reads* spanning the entire gene @TGNieto 

And here's my final one for today - HLA-DPA1/DPB1 - really challenging because of repeats, poor mapping and alignment difficulties. Just look at the two panels. The short reads just don't cut the mustard, but the long reads resolve the entire region, error free, without any probs 

There is so much more data here to explore, and we are currently doing a comparison of all the datasets we are generating with Clair3, PEPPER-Deepvariant, Sniffles, CuteSV and many other nanopore specific tools to see how much more "hidden" information there is in cancer
As a teaser, longer reads seem to be equivalent to SR when processed carefully for CNA - but needs work.
Left - are short reads
Right - is LSK114

Left - are short reads
Right - is LSK114


Many thanks to all the @beggs_lab who make this possible, as well as @JoStockton1 for doing the hard graft in the lab. Also thanks to the team at @uob_bear who provide all the GPU HPC grunt to do these analyses and cheaper than on the cloud (free if you work at @unibirmingham)
See you all at London Calling #NanoporeConf tomorrow!
• • •
Missing some Tweet in this thread? You can try to
force a refresh