
Want to apply #deeplearning in your #regulatory #genomics analyses but overwhelmed by suite of interpretation tools? Check out ExplaiNN: interpretable and transparent neural networks for genomics with @NovakovskyG @ofornes @sara_mostafavi @WyWyWa 🧵👇
biorxiv.org/cgi/content/sh…
biorxiv.org/cgi/content/sh…
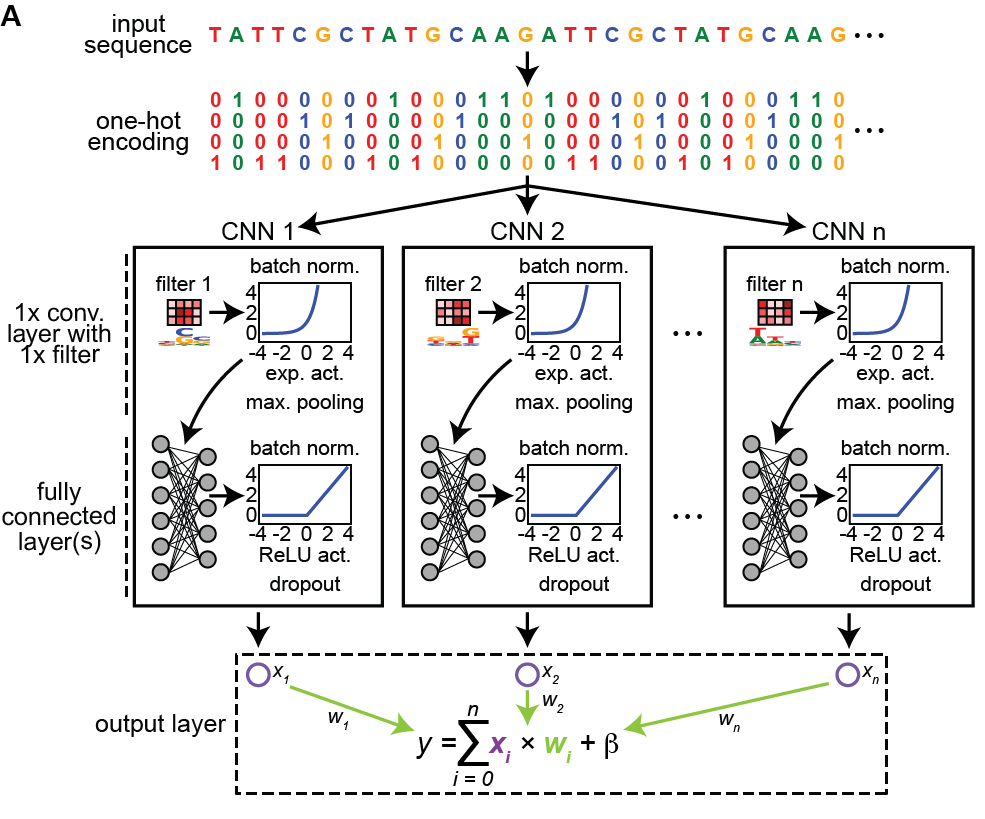
ExplaiNN is an adaptation of NAMs from @agarwl_ & team for genomic sequences - predictions are a linear combination of independent CNNs- each consisting of 1 conv. layer with a single filter and two FC layers. This seemingly simple approach performs similar to multi-layer CNNs1/6 
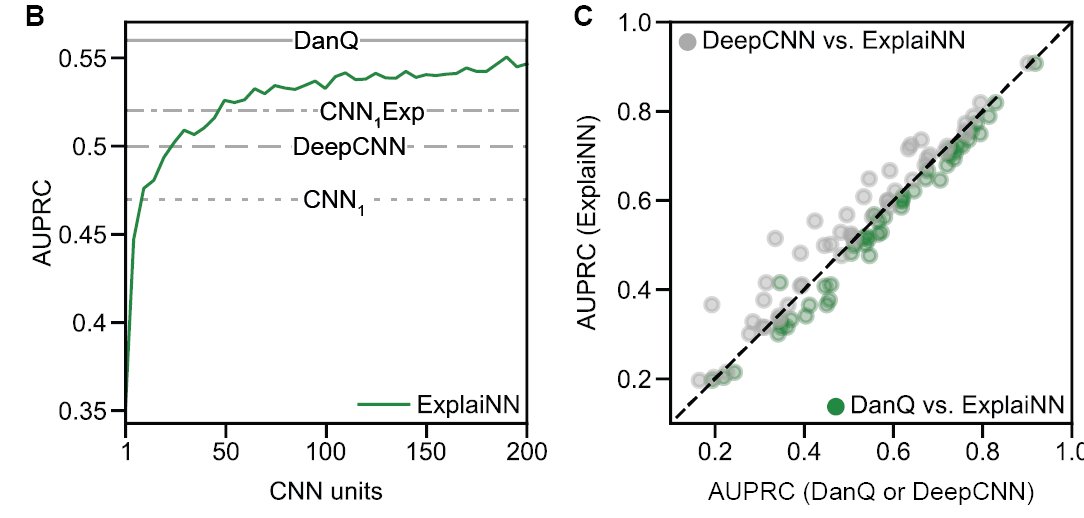
It provides importance of discovered motifs by mere visualisation of weights of the output layer. No need for filter nullification or clustering of importance scores from attribution methods! ExplaiNN on ATAC-seq of mouse immune system recapitulates results of AITAC model 2/6 

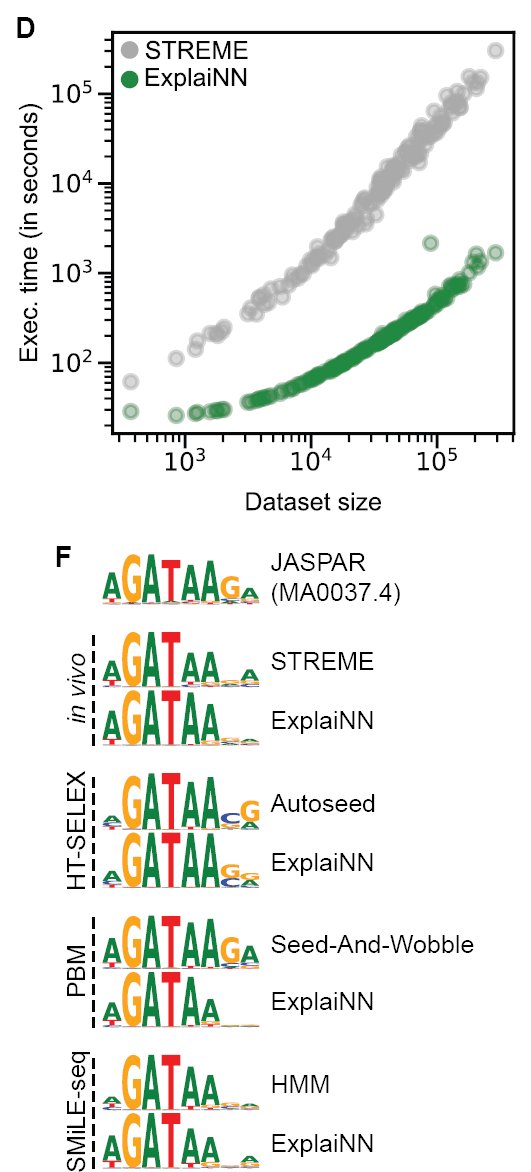
Applied to de novo motif discovery, ExplaiNN detects equivalent motifs to those obtained from specialised algorithms (for e.g. STREME, seed and wobble, Autoseed) across a range of datasets (PBM, HT-SELEX, SMiLE-seq) with orders of magnitude faster run-time 3/6 
Finally,ExplaiNN is a flexible framework to incorp. pre-trained TF binding models/PWMs from JASPAR. No need for post-hoc annotation of conv filters!Using pretrained models also distinguishes between TFs of the same family and generates an interpretable embedding of sequences 4/6 

More results in the paper on sc-ATAC seq for identifying key TFs active in granular cell-types and comparison with DeepSTARR(nature.com/articles/s4158…) for assessing non-linear motif interactions. 5/6
We hope that ExplaiNN will accelerate the adoption of deep learning tools in routine genomic analyses by non-experts as well. The python package and notebooks are available at github.com/wassermanlab/E…
6/6
6/6
On a personal note,this project marks the end of almost 2 years of work with @WyWyWa lab. In particular, it was an immense pleasure to work with my dear friend and colleague @NovakovskyG
Discussing science and life with him at Jericho beach over sunset is one of my fondest memories from Vancouver. @NovakovskyG defends his PhD thesis soon and is looking for ML roles preferably in Biology /Healthcare domain. Do reach reach out to him with relevant opportunities
This thread from @WyWyWa covers some more nuances of the approach
https://twitter.com/WyWyWa/status/1528794559962001408?t=LHMYc3QGXC9fuVvMgnuCMg&s=19
• • •
Missing some Tweet in this thread? You can try to
force a refresh



