
We now have enough co-circulation of Omicron sub-lineages BA.4, BA.5 and BA.2.12.1 in the US to make an assessment of relative fitness between these viruses. 1/12
Pango lineages BA.4 and BA.5, corresponding to @nextstrain clades 22A and 22B, were first described in South Africa by @Tuliodna et al and possess key spike mutations L452R and F486V. 2/12 

Initial spread of Pango lineage BA.2.12.1, corresponding to @nextstrain clade 22C, centered on New York and the Northeast, but has since spread more widely in the US and elsewhere. It possesses key spike mutation L452Q. 3/12 
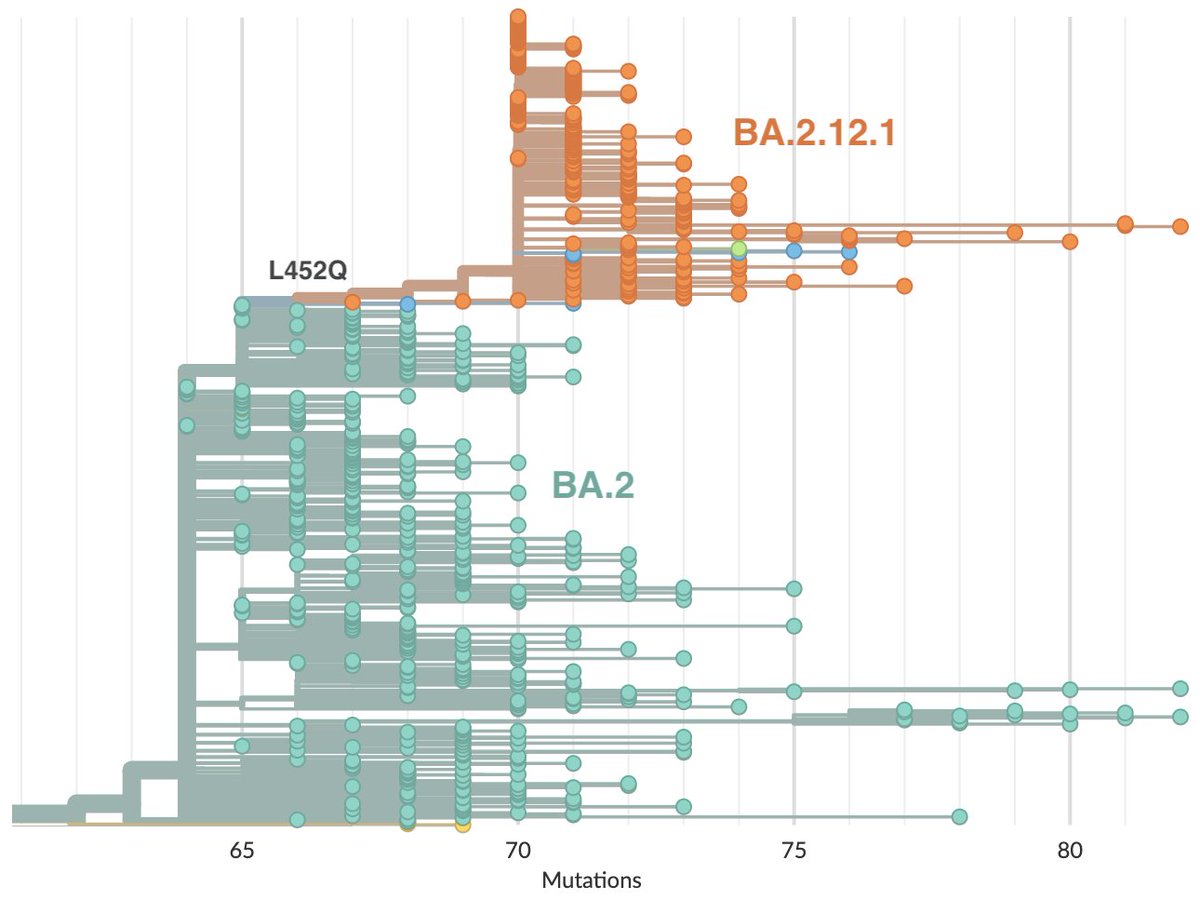
Previously, we could observe relative fitness based on frequency increase in South Africa of BA.4 and BA.5 and separately, relative fitness in the US of BA.2.12.1. But comparisons across geographies of frequency growth rate are not fully reliable. 4/12
https://twitter.com/trvrb/status/1516147488587075584
Ideally, we want to compare frequency growth rate in the same geography. Focusing on the US, we see that BA.2.12.1 currently has a logistic growth rate of 0.05 per day, while BA.4 and BA.5 have logistic growth rates of 0.09 and 0.14 per day. 5/12 
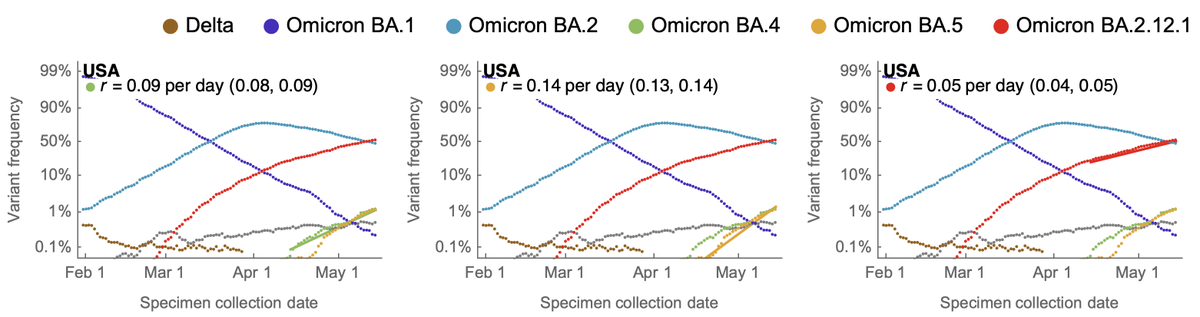
This suggests BA.4 and BA.5 are fitter than BA.2.12.1 as one might expect given the additional spike F486V mutation (plus mutations elsewhere in the genome), and that we expect BA.4 and BA.5 to continue to increase in frequency on the background of BA.2 and BA.2.12.1. 6/12
We can assess variant-specific Rt using method developed by @marlinfiggins (bedford.io/papers/figgins…), where we observe current Rt for BA.2.12.1 of ~1.0, compared to Rt of BA.4 of ~1.2 and BA.5 of ~1.5. 7/12 
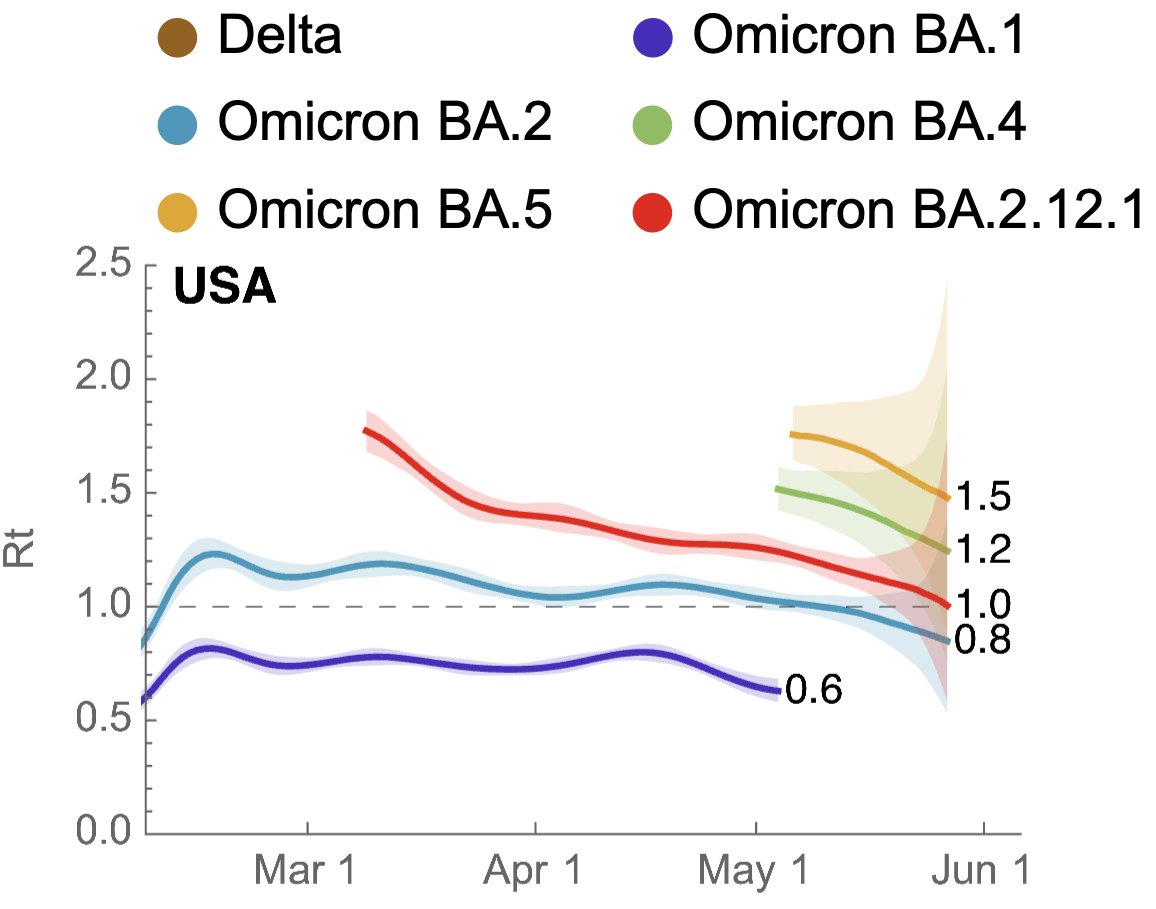
This along with current frequency of BA.2.12.1 indicates that rise in current case counts can be at least in part attributed to BA.2.12.1 epidemic and we additionally see growing epidemics of BA.4 and BA.5 in absolute case counts. 8/12 
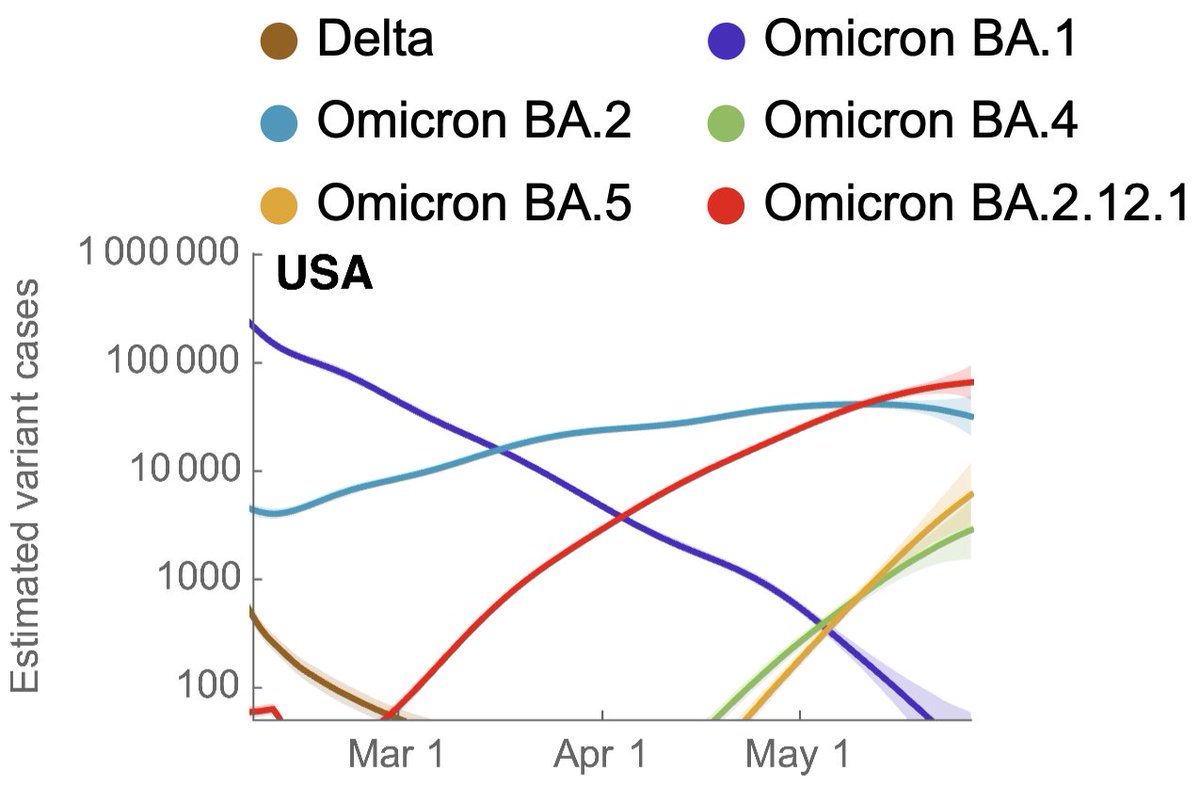
This pattern is mirrored across other countries where we see growing epidemics of BA.2.12.1, BA.4 and BA.5, particularly in Portugal where BA.5 got an early start and is beginning to push cases past BA.2 levels. 9/12 
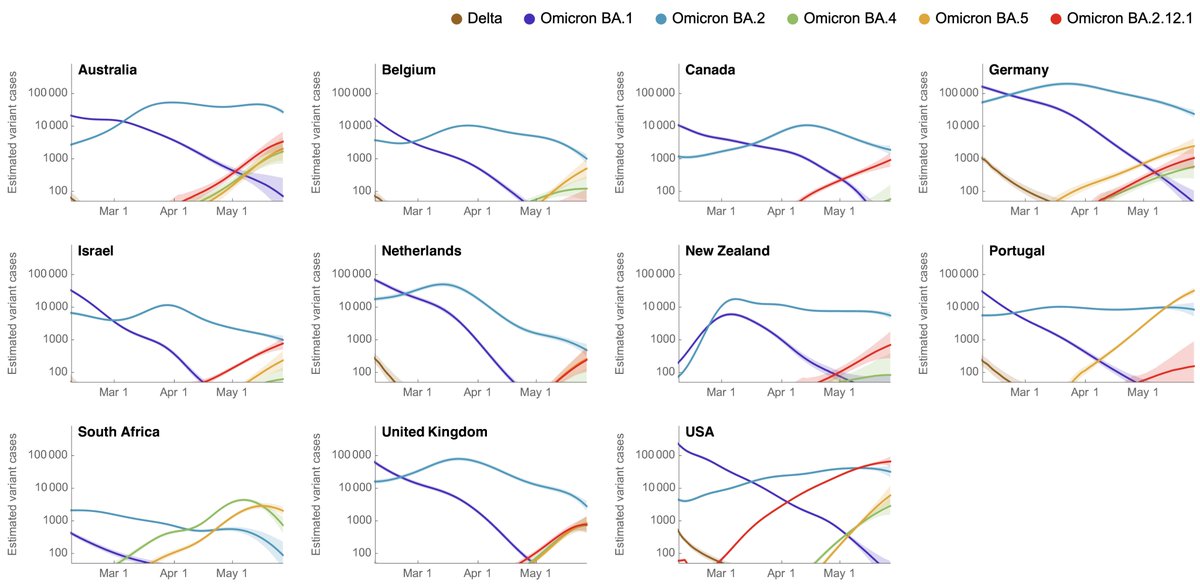
More detailed plotting of variant circulation across countries is viewable at github.com/blab/rt-from-f… and across states is viewable at github.com/blab/rt-from-f…. 10/12
There is a notable drop in neutralization titer against BA.4 and BA.5 viruses in boosted individuals and individuals with BA.1 breakthrough infections (figure from Qian Wang, David Ho and colleagues biorxiv.org/content/10.110…). 11/12 
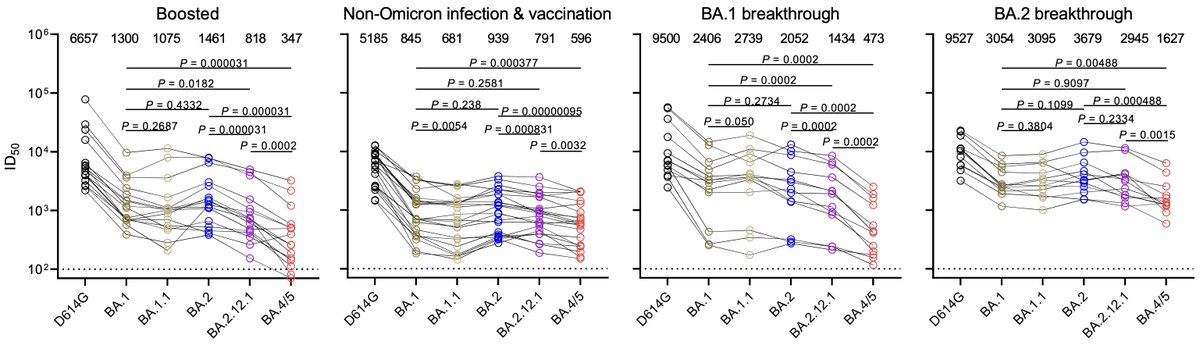
Thus, we expect at least some portion of the BA.4 / BA.5 epidemics to be driven by increased vaccine breakthrough and increased reinfection relative to current BA.2 circulation. 12/12
• • •
Missing some Tweet in this thread? You can try to
force a refresh












