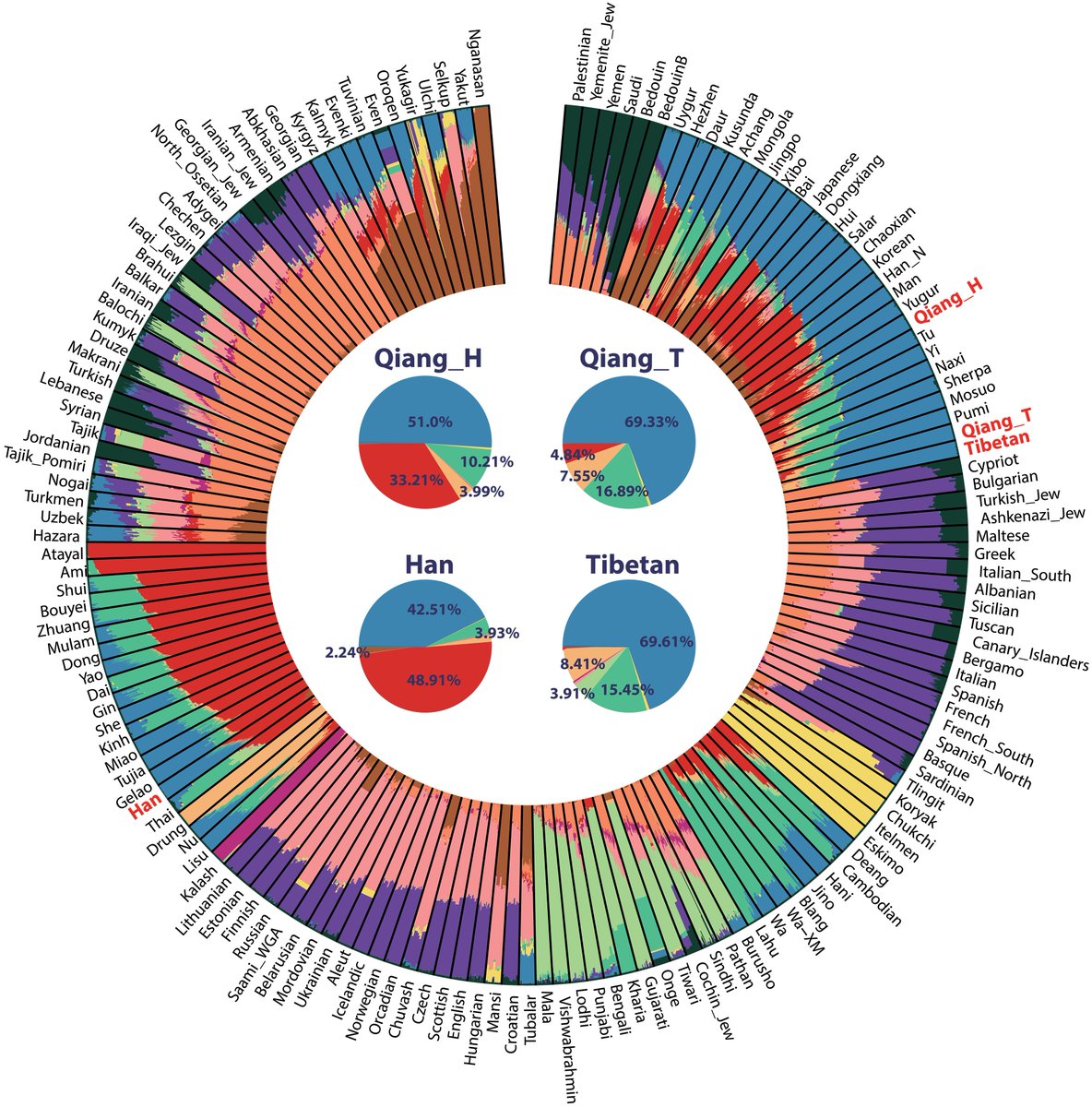
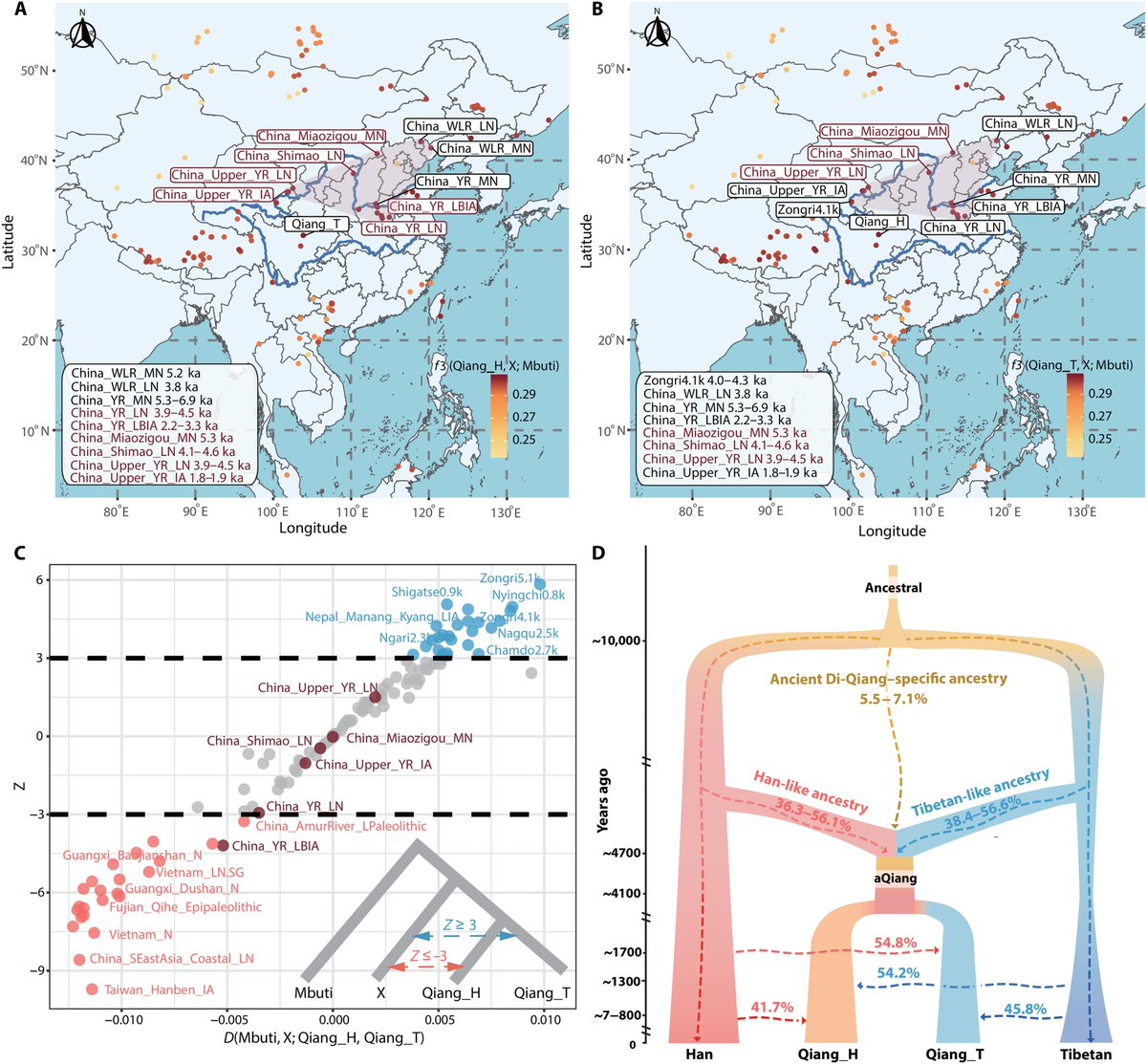
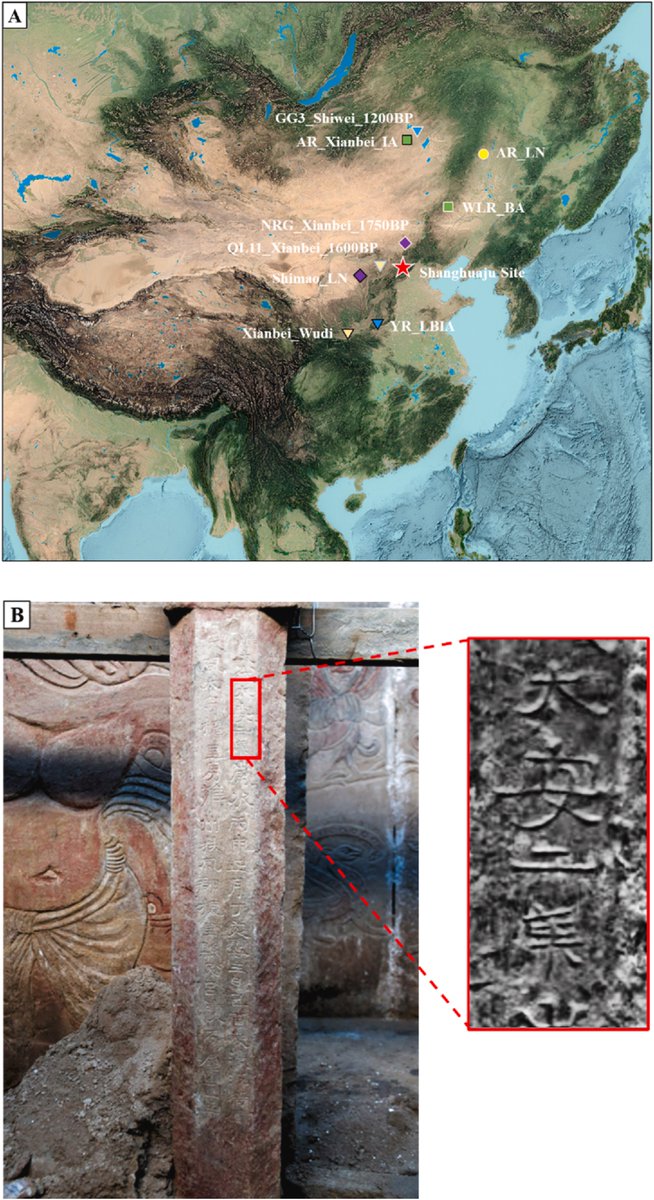
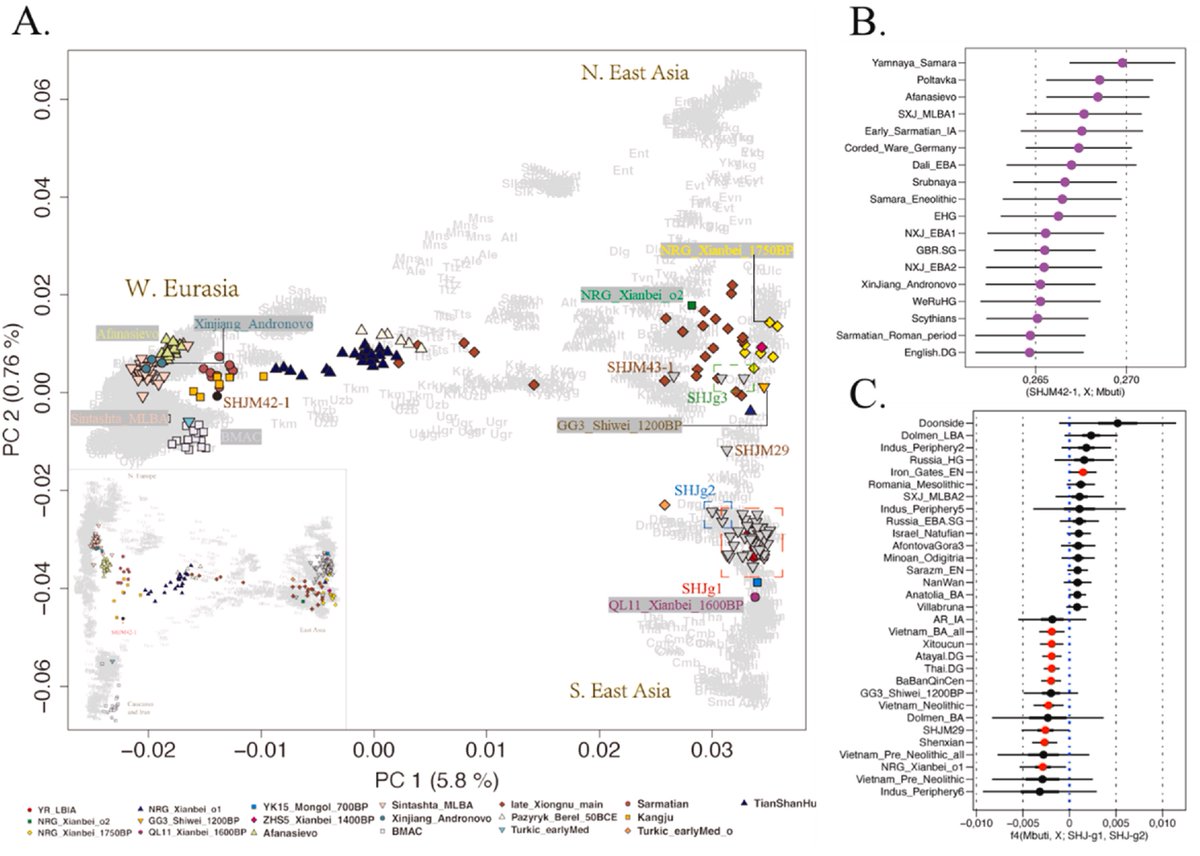
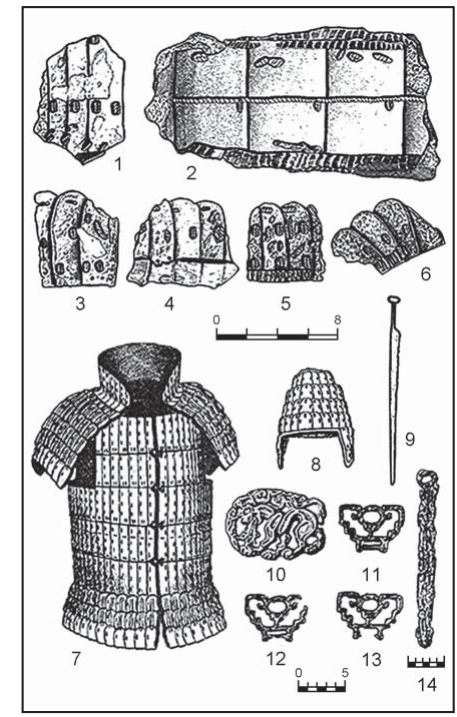
Comparison between G25 and Fst as way of determining genetic distances for Yamnaya_Samara between ancient and modern populations.
1. Fst.
1. Fst.

Unroll @threadreaderapp
• • •
Missing some Tweet in this thread? You can try to
force a refresh