
Excited to share our new preprint describing
• how to achieve resistance to all viruses,
• a novel technology that biocontains engineered genetic information, &
• how viruses overcome genetic-code-based #virus resistance
biorxiv.org/content/10.110…
Tweetorial 1/n
• how to achieve resistance to all viruses,
• a novel technology that biocontains engineered genetic information, &
• how viruses overcome genetic-code-based #virus resistance
biorxiv.org/content/10.110…
Tweetorial 1/n
This work was an incredible collaborative effort: with @SvenjaVinke, Regan Flynn, @geochurch,
Huge thank you to Michael @baym @implosian @EllieRand3,
and @BogdanBudnik @wyssinstitute & @GenScript
Special thanks to @behnoush_h for graphics help, and all co-authors!
2/n
Huge thank you to Michael @baym @implosian @EllieRand3,
and @BogdanBudnik @wyssinstitute & @GenScript
Special thanks to @behnoush_h for graphics help, and all co-authors!
2/n
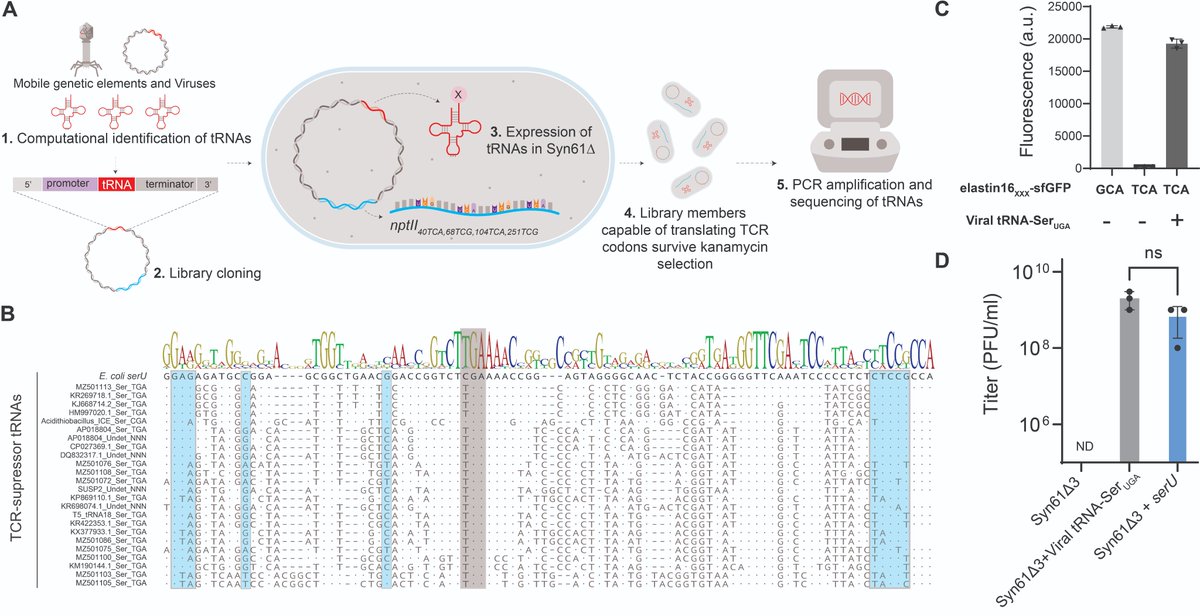
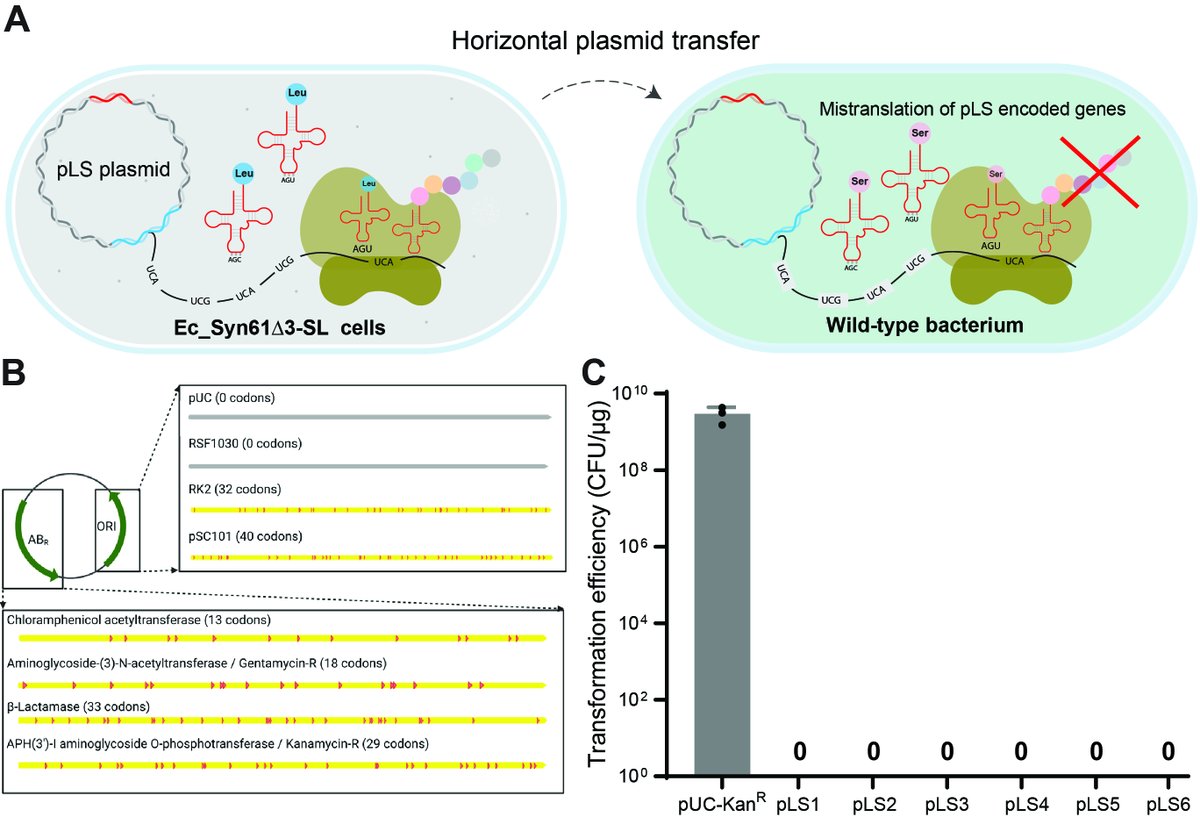
We show that the horizontal transfer of tRNA genes rapidly abolishes the genetic code-based isolation of recoded organisms and rescues viral replication in Syn61Δ3.
We also detected the presence of mobile tRNA genes in numerous viral genomes and mobile genetic elements.
3/n
We also detected the presence of mobile tRNA genes in numerous viral genomes and mobile genetic elements.
3/n
Next, in collaboration with Michael @baym @implosian @EllieRand3, we isolated 12 lytic bacteriophages from environmental samples that lyse Syn61Δ3 and form plaques on this recoded E. coli strain, despite the genome-wide lack of 3 codons and 2 serine tRNAs + Release Factor 1
4/n
4/n
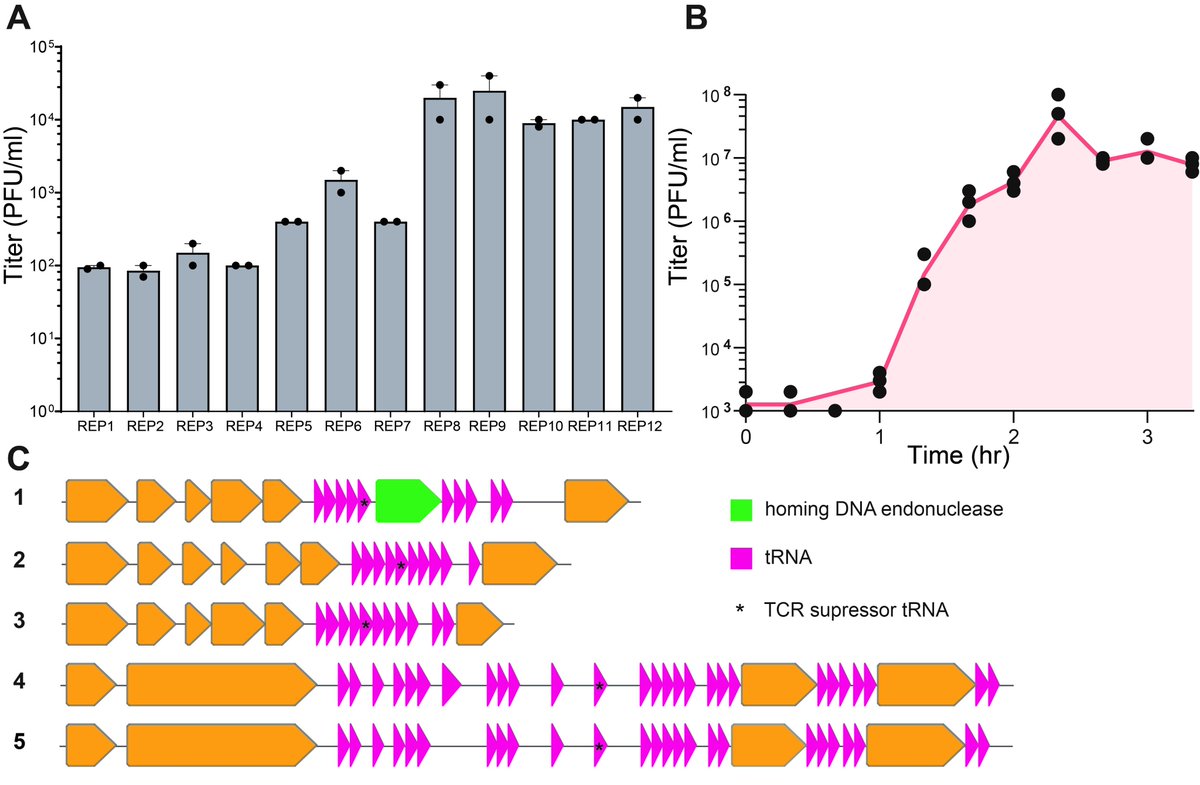
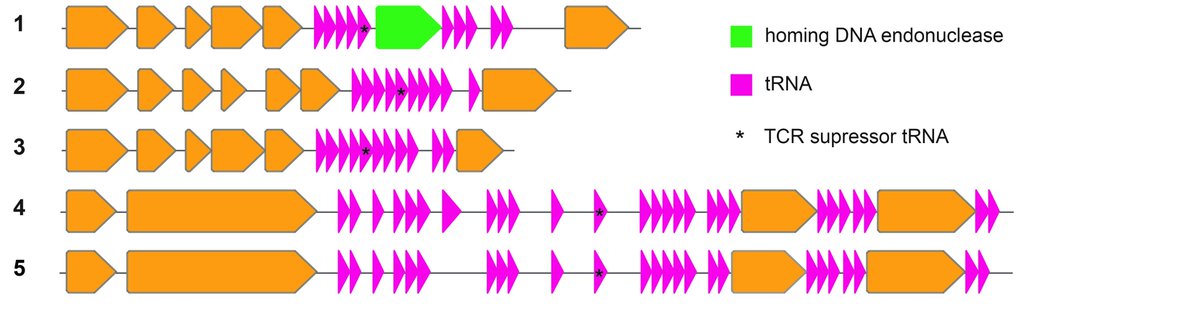
These REP (Recoded E. coli phages) carry and express up to 27 tRNA genes, including a serine tRNA with a UGA anticodon that substitutes the missing serU and serT tRNAs of Syn61Δ3. 
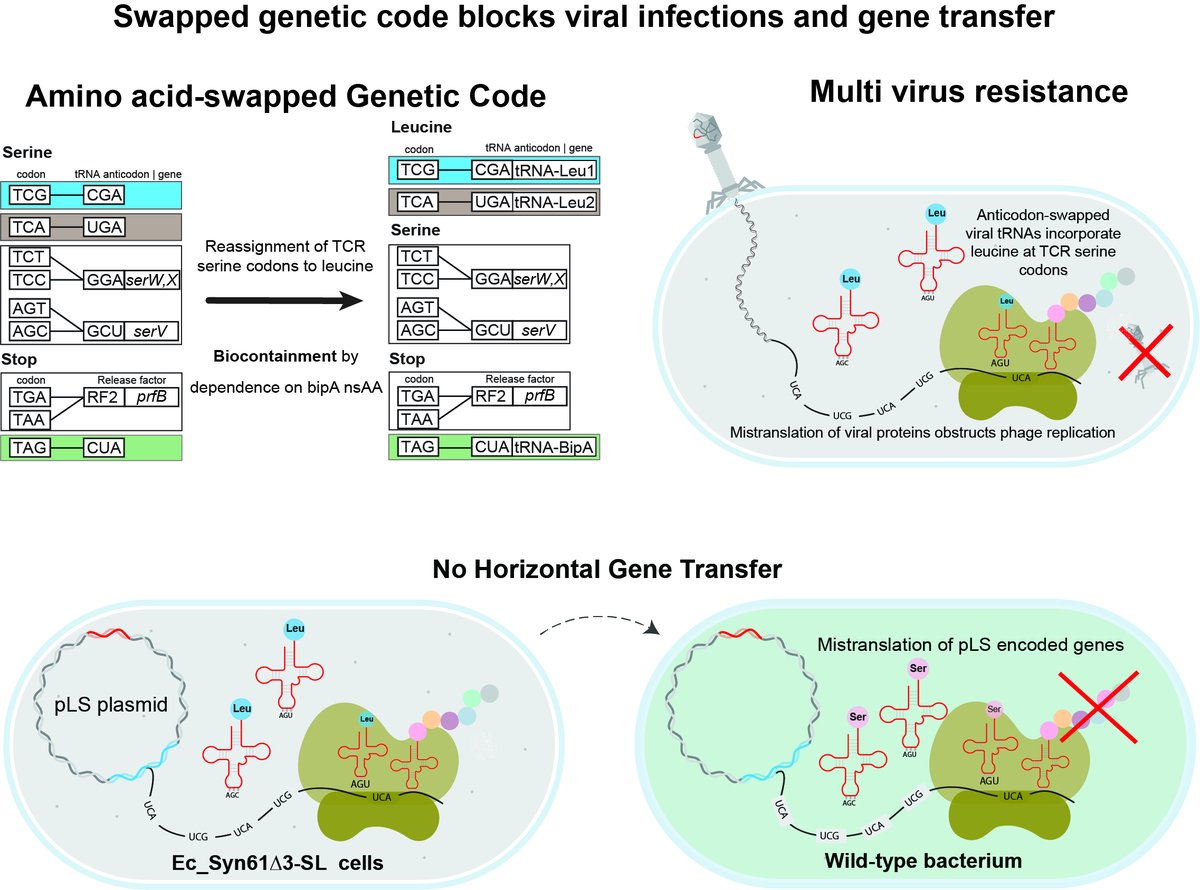
By repurposing viral tRNAs, we then develop a recoded organism with an artificial, amino-acid-swapped genetic code that resists all tested viruses.
This swapped genetic code renders cells completely resistant to viral infections, incl. viruses in sewage & our 12 REP phages.
This swapped genetic code renders cells completely resistant to viral infections, incl. viruses in sewage & our 12 REP phages.
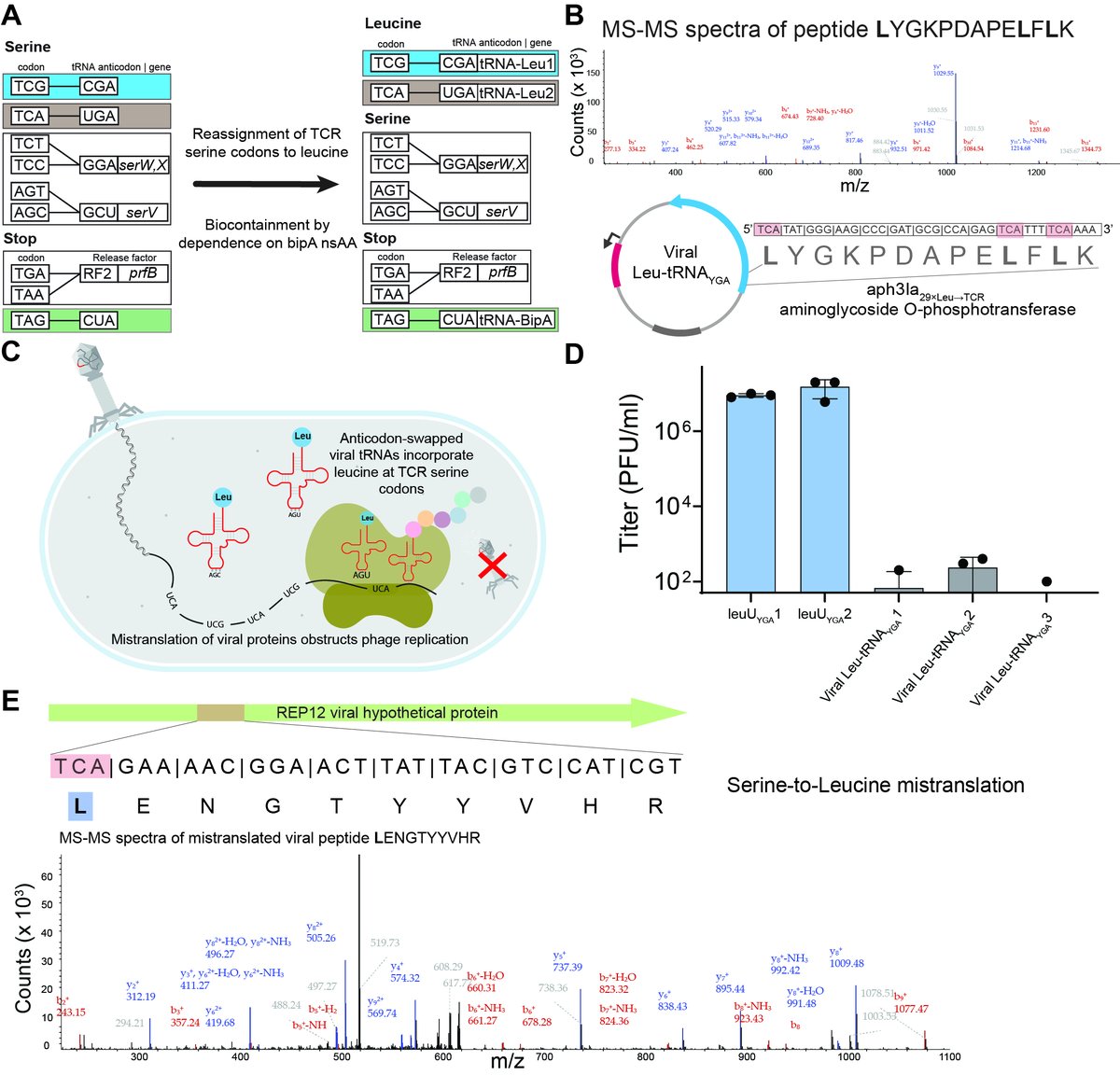
This amino-acid-swapped genetic code also serves as a firewall that prevents the escape of engineered genetic information from GMOs -- by using serine codons to produce leucine-requiring proteins.
7/n
7/n
Finally, we also repurpose the third free codon in Syn61Δ3 to biocontain this virus-resistant host via dependence on an amino acid not found in nature (bipA).
We are also thankful to Jason W. Chin and his team @MRC_LMB for their pioneering work with Syn61Δ3 and for sharing the strain via @Addgene #OpenScience
9/n
9/n
• • •
Missing some Tweet in this thread? You can try to
force a refresh



