
PAPER OUT !! 🎉🥂🍾 @nature
nature.com/articles/s4158…
Viral restrictor APOBEC3A is a major cause of mutational signatures found in >50% of cancers + more, Tweetorial👇
With @J_Maciejowski
Via WGS/251 cancer clones (available!)
@broadinstitute
@MSKCancerCenter
@sangerinstitute
nature.com/articles/s4158…
Viral restrictor APOBEC3A is a major cause of mutational signatures found in >50% of cancers + more, Tweetorial👇
With @J_Maciejowski
Via WGS/251 cancer clones (available!)
@broadinstitute
@MSKCancerCenter
@sangerinstitute

The APOBEC family of cytosine deaminases has been implicated as a source of some of the most prevalent mutational signatures in cancer. BUT, causal links between endogenous APOBECs and mutations in cancer haven’t been established, leaving the mechanisms poorly understood. 
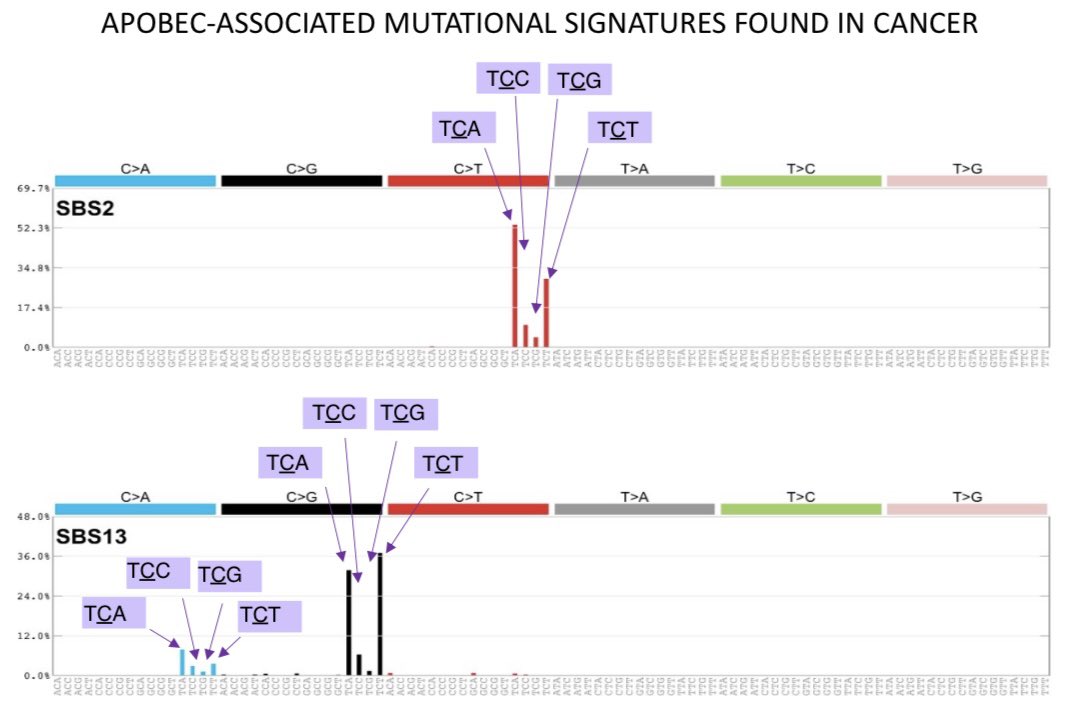
To investigate the mechanisms underlying acquisition of APOBEC-associated mutational signatures in cancer, we deleted implicated genes from human cancer cell lines that we previously showed generate the relevant signatures naturally over time (cell.com/cell/pdf/S0092…) 

Analysis of non-clustered and clustered signatures, acquired over controlled timeframes in vitro, across WGS of 251 breast, bladder and lymphoma cancer cell line clones revealed that APOBEC3A deletion diminished majority of the APOBEC3-associated mutational signatures, eg: 

Cytosine mutations induced by APOBEC3A in human cancer cell lines are enriched at YTC trinucleotides context (Y=pyrimidine), most prevalently found in cancers with evidence of exposure to APOBEC-associated mutagenesis (Chan et al, NatGen, 2015 @DmitryYeast)
In all examined cell lines, protein levels of APOBEC3A (new antibody by @ALLIEgator__ @maciejowskilab !) were minimal relative to APOBEC3B- an observation prevalently found in cancer and previously (and continuously!) used to nominate APOBEC3B as the major mutator in cancer. Eg: 
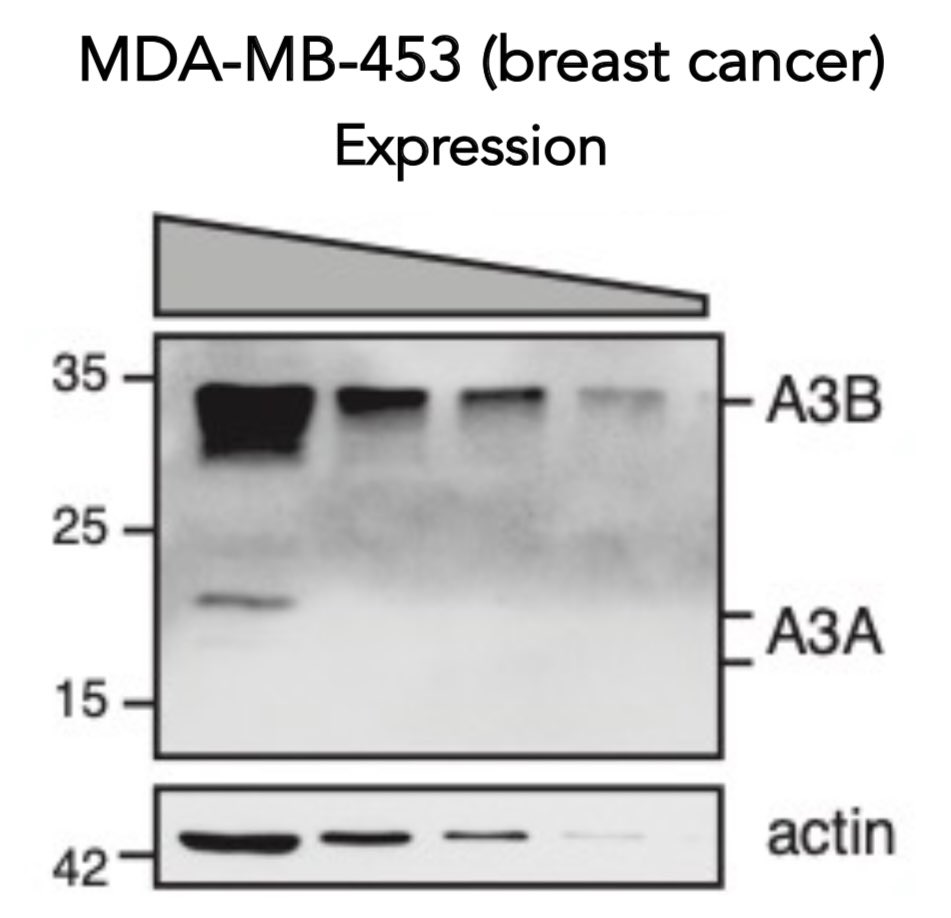
APOBEC3B contributes smaller mutation burdens, while it can (!) sometimes restrain APOBEC3A-dependent mutagenesis, mechanism unknown (on it!).
Note:APOBEC3A/APOBEC3B KOs continue (!) to acquire small burdens of the relevant signatures – another APOBEC matters (??)
Eg:
Note:APOBEC3A/APOBEC3B KOs continue (!) to acquire small burdens of the relevant signatures – another APOBEC matters (??)
Eg:
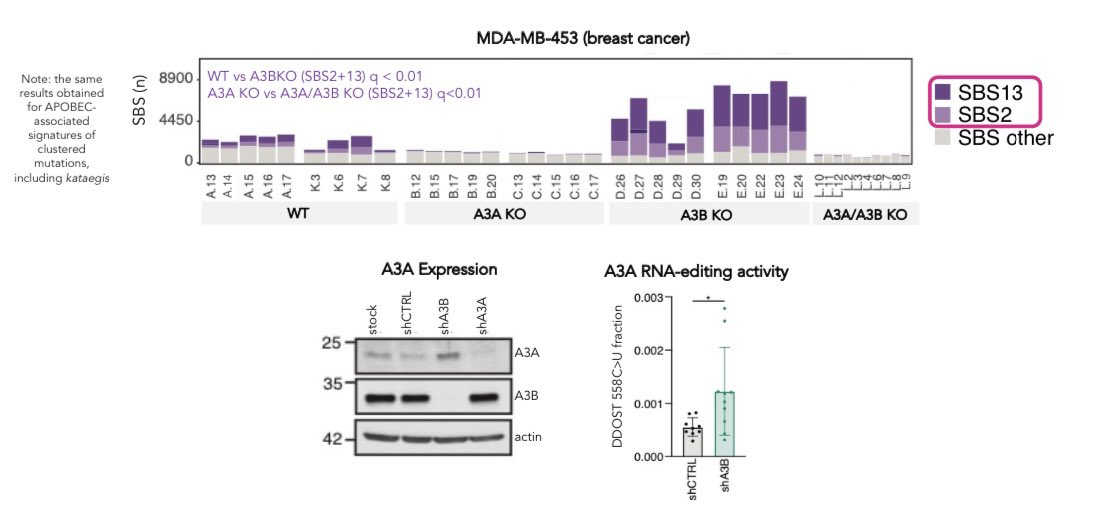
Cancers are usually defined as APOBEC3A-like or APOBEC3B-like. Our data shows that multiple APOBEC enzymes, including APOBEC3A and APOBEC3B, can contribute mutations in a single cancer.
APOBEC3B may be a more relevant mutator in a smaller proportion of cancers with APOBEC-associated mutations enriched in APOBEC3B-preferred (@DmitryYeast) RTC (R= purine) sequence contexts
Uracil glycosylase UNG and translesion polymerase REV1 are the relevant mediators of APOBEC-induced edits: UNG is required for APOBEC-mediated transversions, whereas REV1 mediates generation of most APOBEC-associated mutations. Eg: 
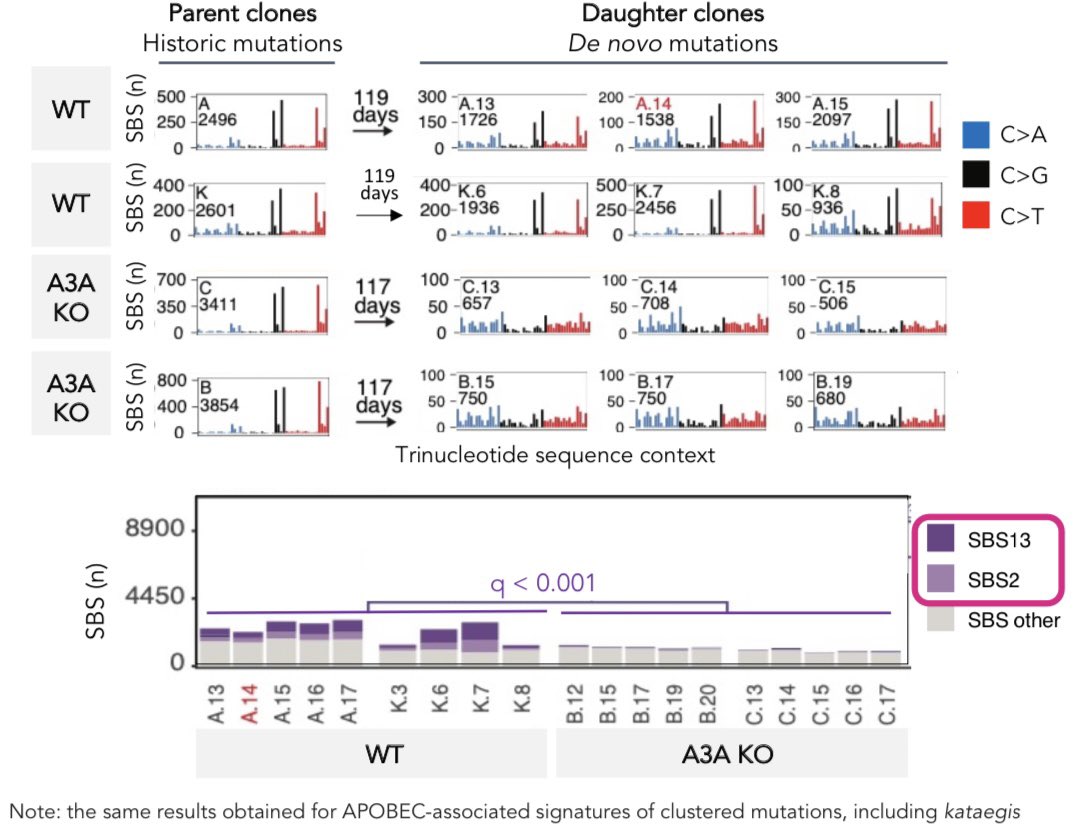
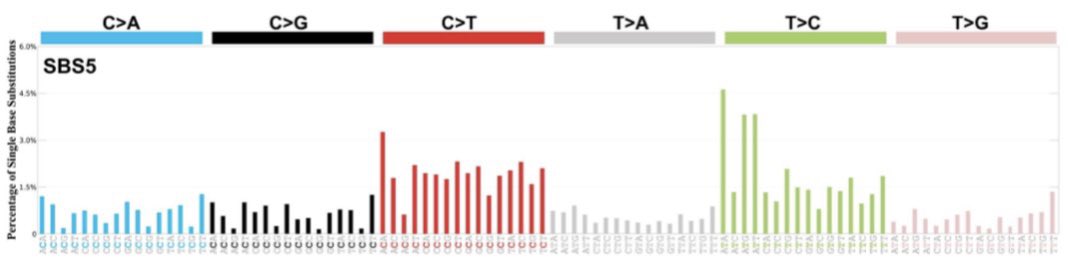
Along the way, we stumbled upon REV1 being the possible origin of ‘mutational signature 5’, found in all human genomes (cancer and non-malignant) analyzed to date, thus representing an imprint of one of the most prevalent mutational processes in humans as such. 
Critical contributions @ALLIEgator_ @kevanchu @yanyangch; #MikeStratton’s lab; also @ErikNBergstrom #Alexandrov’s lab @UCSD; @MRC_LMB. Support 🙏@CancerGrand @EMBO @EMBOFellows @theNCI @pewtrusts @TheVFoundation @wellcometrust @CR_UK. Graphics thx to @GraphicAnja @patternvizlab
• • •
Missing some Tweet in this thread? You can try to
force a refresh



