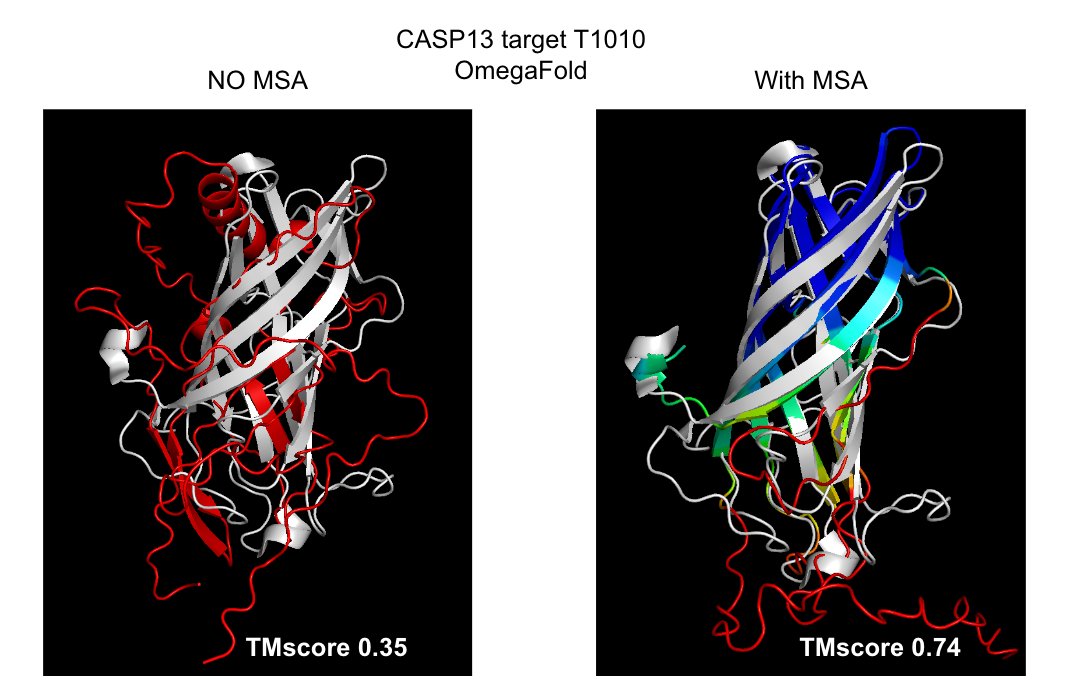
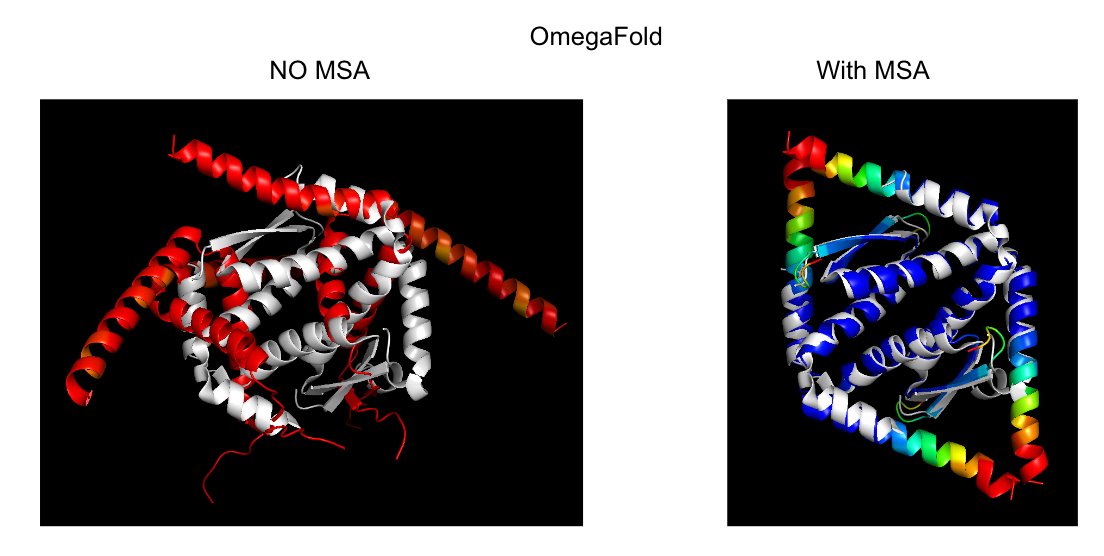
Found an example from @Alexis_Verger that OmegaFold fails on (and has low confidence). But if you hack OmegaFold to use an MSA input, it gets it right (and has high confidence)! 

https://twitter.com/Alexis_Verger/status/1555134615328899072


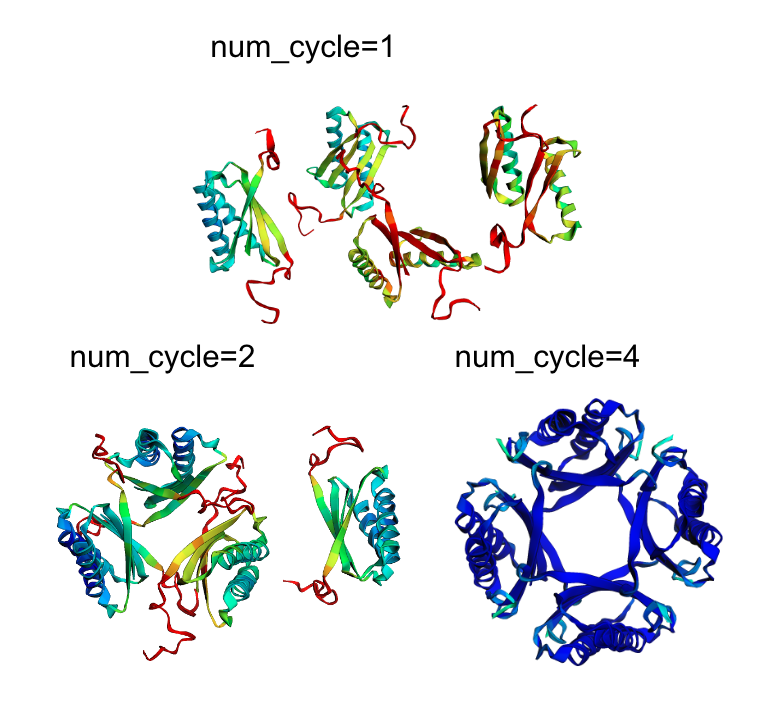
@Alexis_Verger another example: a homo-tetramer that fails with OmegaFold. But if you add an MSA, it works! 🙃

https://twitter.com/onoda_hiroki/status/1554918403172798464
@Alexis_Verger Experimental notebook, if anyone wants to try! 😎
colab.research.google.com/github/sokrypt…
colab.research.google.com/github/sokrypt…
• • •
Missing some Tweet in this thread? You can try to
force a refresh