
Scientist, Assistant Professor @MITBiology, #FirstGen, ProteinBERTologist, 🇺🇦
No Human is illegal.
Moving to: https://t.co/sow6IRD3jj
How to get URL link on X (Twitter) App


 Traditional methods approx this signal by taking a multiple sequence alignment of a protein family and computing the inverse covariance matrix. For pLMs we extract it by computing a jacobian over the sequence track (for esm3, structure is masked). (2/8)
Traditional methods approx this signal by taking a multiple sequence alignment of a protein family and computing the inverse covariance matrix. For pLMs we extract it by computing a jacobian over the sequence track (for esm3, structure is masked). (2/8)https://x.com/sokrypton/status/1752914756250259608

https://twitter.com/ProteinBoston/status/1802093052216869104For context, traditional methods like GREMLIN extract coevolution from input MSA. If you make the assumption that data is non-categorical, you can approximate the coevolution signal via inverse-covariance matrix (2/9). arxiv.org/abs/1906.02598


 We believe AlphaFold has learned some approximation of an "energy function" and a limited ability to explore. But this is often not enough to find the correct conformation, and often an MSA is required to reduce the search space. (2/9)
We believe AlphaFold has learned some approximation of an "energy function" and a limited ability to explore. But this is often not enough to find the correct conformation, and often an MSA is required to reduce the search space. (2/9) 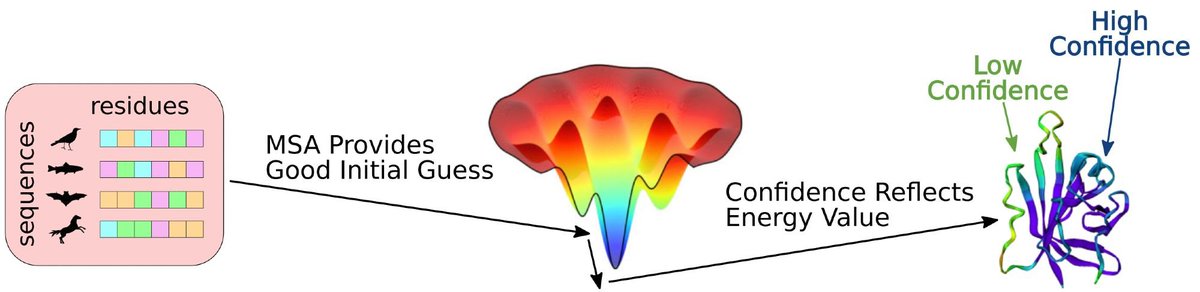
https://twitter.com/hendrik_dietz/status/1629749217399808001



https://twitter.com/MartinPacesa/status/1601511762356228096Though Zscores can be a little misleading. If everyone is the same on average, and one group does really well on a particular target compared to everyone else, they HUGE boost. Here are the same data (same order, but plotting average GDT_TS). (2/6)






https://twitter.com/Alexis_Verger/status/1555134615328899072

 @Alexis_Verger
@Alexis_Verger 

https://twitter.com/peng_illinois/status/1554831291794657282
 In the spirit of hacking methods for what they were not trained to do. I added support for chain breaks using the residue index offset trick! 😇
In the spirit of hacking methods for what they were not trained to do. I added support for chain breaks using the residue index offset trick! 😇 

 Here is the Pseudo-code of the changes we made and the effects on compile time. (2/2)
Here is the Pseudo-code of the changes we made and the effects on compile time. (2/2) 
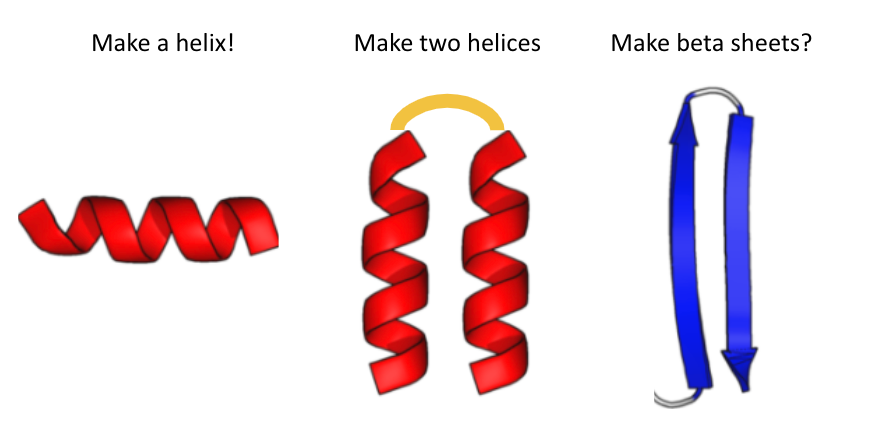
 I gave them the following cheat sheet: 😅 (2/5)
I gave them the following cheat sheet: 😅 (2/5) 

