
Just released another #Python package for #bioinformatics: #Cython bindings to FAMSA, a very fast algorithm for #MultipleSequenceAlignment by @sdeorowicz, Agnieszka Debudaj-Grabysz and @AdamGudys! Get it here: github.com/althonos/pyfam…
Just create an Aligner object (with additional configuration if you want), then give it some sequences to align: 
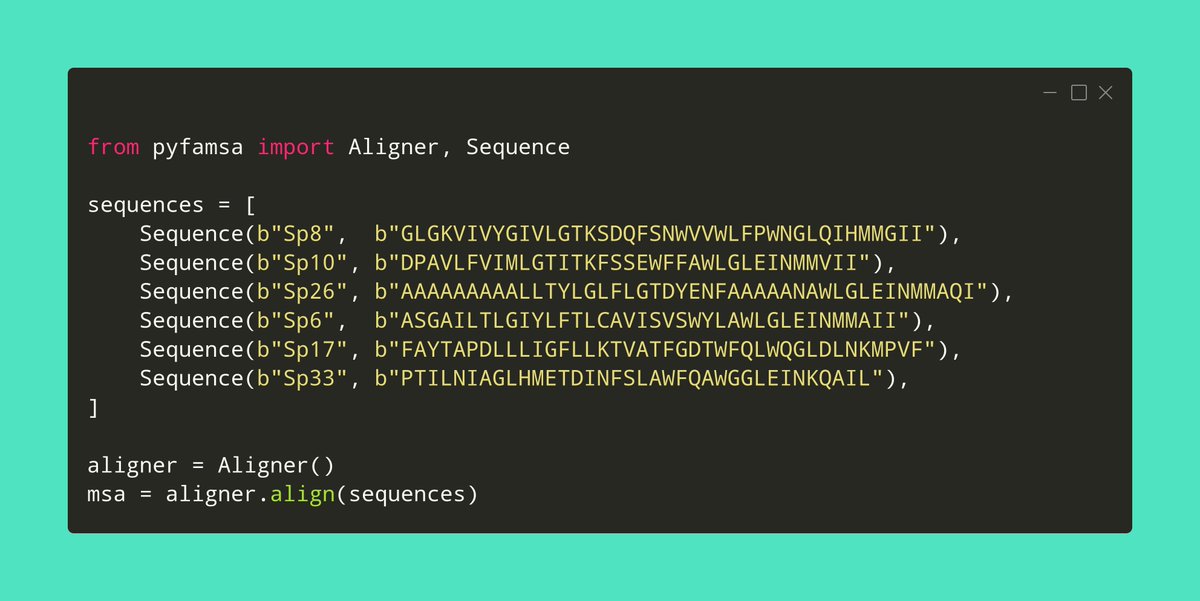
It's missing some small things that I'll add over time, but for now the key features are there, and you could use it in your Python or #snakemake workflows in place of the FAMSA binary.
And after you're done with the alignment, you can always trim it with PytrimAl, all inside a Python interpreter!
https://twitter.com/althonos/status/1532481359016611855
• • •
Missing some Tweet in this thread? You can try to
force a refresh



