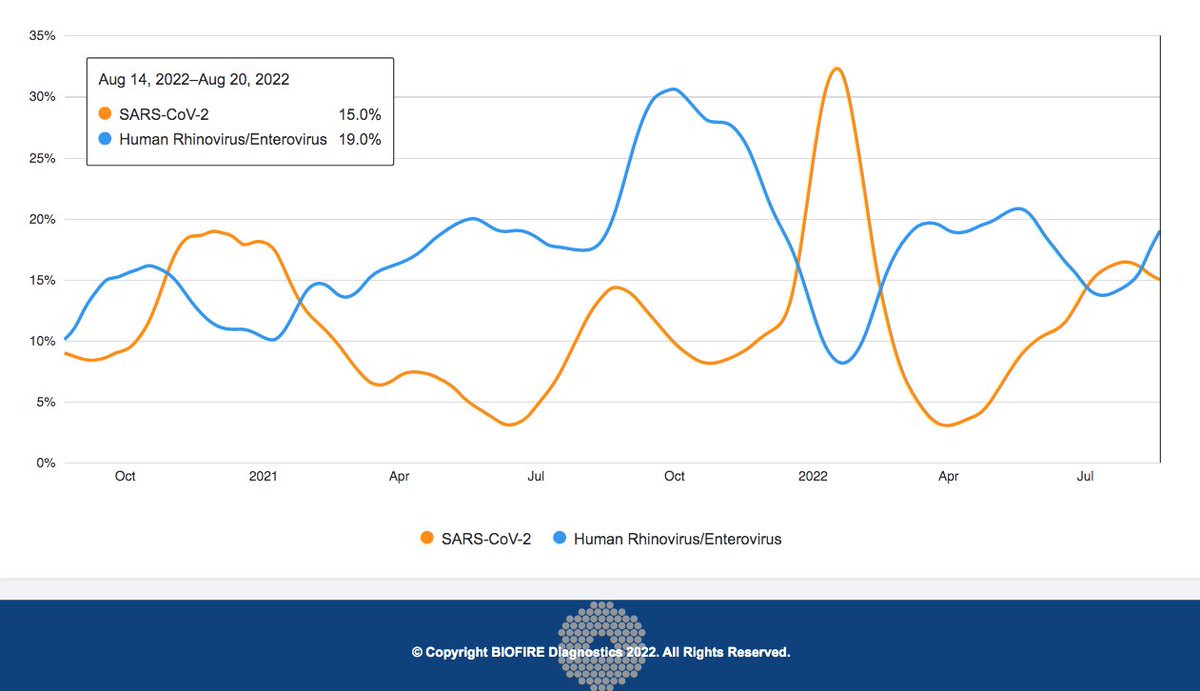
Inverse relationship between rhinovirus & SARS-CoV-2 still holding strong. Seems clear some form of viral interference is happening here. But is the causation all one way (SARS2-->RV) or does it go both ways? As someone likely suffering an RV infection right now, I'm curious 1/10 
There is some evidence that rhinovirus infection protects against Covid by activating interferon-stimulated genes (ISG). ISG are activated upon SARS-CoV-2 infection as well, but with a delay that allows the virus to replicate & establish infection. 2/10 rupress.org/jem/article-st… 

Here's another 2021 study showing experimental evidence that RV impedes Covid. Remarkably, SARS-CoV-2 was inhibited even when rhinovirus was introduced 24 hours after inoculation. 3/10 academic.oup.com/jid/article/22… 

By blocking the interferon response stimulated by RV infection, the experimenters proved that it was through the stimulating the innate interferon response that RV interferes with SARS-CoV-2. 4/10 

At least one eminent virologist and rhinovirus specialist maintains the viral interference between SARS-CoV-2 and rhinovirus works both ways, as one would expect, even if the interferon response to Covid is somewhat weaker than that evoked by RVs. 5/10
https://twitter.com/MackayIM/status/1380298385920512001
It seems to be generally accepted that a large rhinovirus wave that accompanied the start of the school year in some European countries in 2009 delayed the onset of the H1N1 pandemic flu ("swine flu") waves there. 6/10 

More recent research seems to confirm what was suspected at the time: rhinovirus infection provides protection against influenza A. 7/10 
https://twitter.com/EllenFoxman/status/1302216013358956544

BTW, I'm not trying to make a point here, certainly not that we should seek out RV infection to prevent flu or Covid—RV is making me quite miserable right now. I want to avoid future RV infection as much as possible. I just find this all fascinating. 8/10
Viral interference happens with other viruses as well, and the original Omicron wave drove down the prevalence of almost all other respiratory viruses with one very strange exception: the seasonal coronaviruses. I still don't understand that one. 9/10
https://twitter.com/LongDesertTrain/status/1485967125034508289
Usual caveat: I'm not an expert in these matters and have likely left out important considerations or said something stupid. Corrections & additional information from genuine experts (& others) welcome. 10/10
• • •
Missing some Tweet in this thread? You can try to
force a refresh