
Examples from our Protein Common Assembly Db now on @biorxiv_bioinfo with @QifangXu. Viable assemblies for all Xray structures (EPPIC, @josemduarte), clustered w/ cryoEM, NMR assemblies by Pfam architectures + stoichiometry+ symmetry + interfaces. @buildmodels @PDBeurope @pdbj_en 
https://twitter.com/biorxiv_bioinfo/status/1559386086094381058
We count "Crystal forms (CFs)" - unique crystal unit cells or cryo-EM or NMR experiments for each cluster, and number of uniprot sequences across each cluster. More experimental data --> more support for relevance of biological assembly. Also #PDB's bioassemblies, PISA, EPPIC.
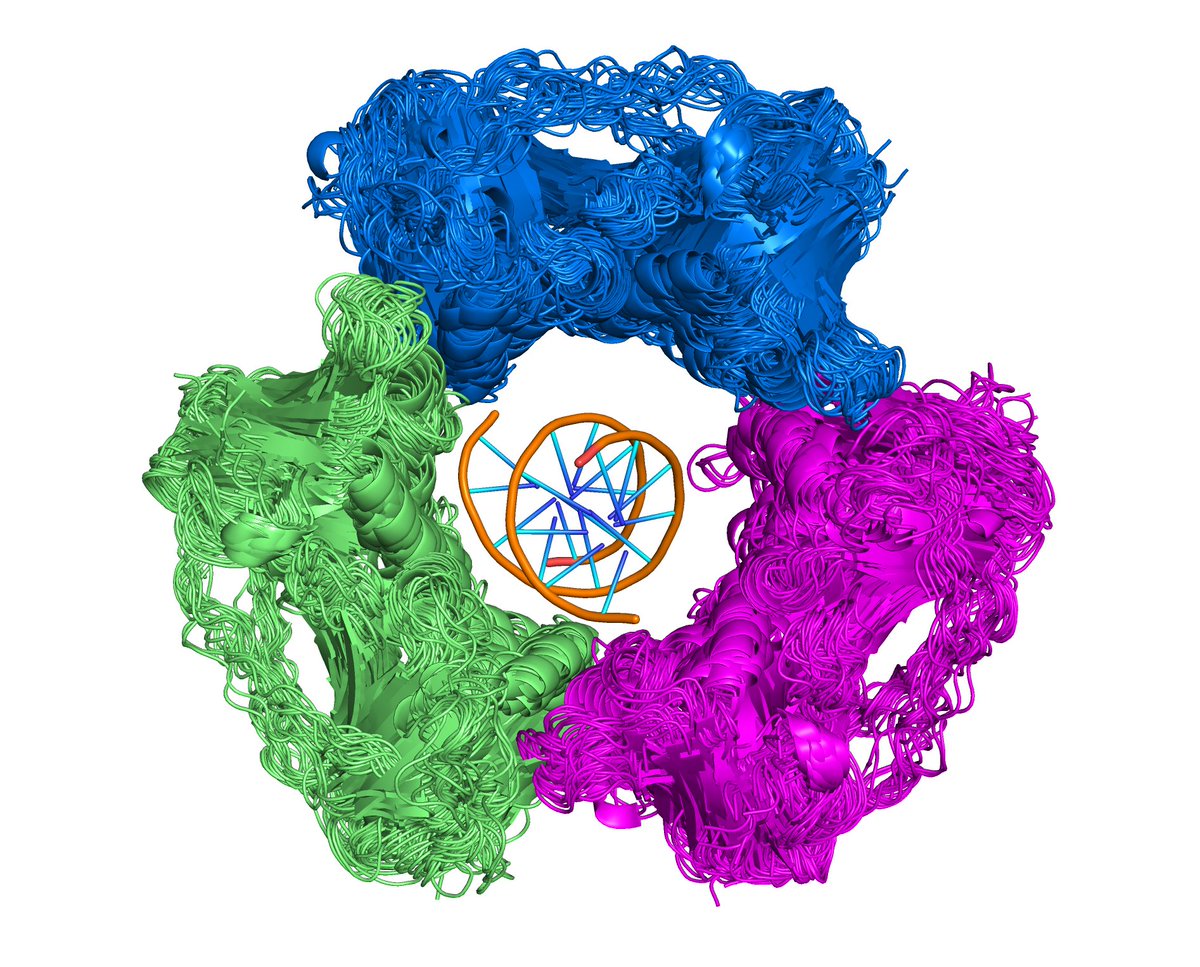
PCNA example from 1st tweet. Largest cluster (#8) of Pfam arch (PCNA_N)_(PCNA_C) has 28 crystal forms (CFs) out of 36 CFs for this Pfam architecture in whole in PDB (78%). 19 UNPs of 23 in whole PDB (83%). 55 entries of 64 in whole PDB (86%).
dunbrack2.fccc.edu/ProtCAD/Result…
dunbrack2.fccc.edu/ProtCAD/Result…

Of 31 CFs (#CFs_UNParch) for these Uniprots in whole PDB, 28 (#CFs_UNPClus) have this assembly (0.9 or 90%). Of 55 entries in cluster, 47 have this assembly in PDB's biol. assemblies (85%). EPPIC and PISA have 32 (58%). Click [8] button in 1st column -> Pymol script & cif files.
This well known assembly is present in some crystals but actually missing in PDB biological assemblies. Example. 
Here are other examples. MHC Class II alpha/beta chains with peptide. Find by searching for Pfam (MHC_II_alpha) or Uniprot (DRA_HUMAN) or PDB (1DLH). 53 CFs, 30 UNPs, 69 entries, 67 PDBBA. dunbrack2.fccc.edu/ProtCAD/Result… 

14-3-3 proteins -- homodimer with peptides. 56 CFs, 11 UNPs, 297 entries. We cluster Pfam architectures with and without peptides separately. dunbrack2.fccc.edu/ProtCAD/Result… 

P53 and P73 tetramers with DNA. 10 CFs, 19 PDB entries. dunbrack2.fccc.edu/ProtCAD/Result… 

CD1 proteins (Pfam MHC_I_3) with T-cell receptors. Two clusters. Cluster 1: alpha-yellow; beta-cyan), 11 CF, 65 entries, all CD1D_MOUSE or HUMAN. Cluster 2 (alpha-orange, beta-blue TCRs), 8 CFs, CD1A, CD1B, CD1C, CD1D. dunbrack2.fccc.edu/ProtCAD/Result… 

FGFR and FGF heterotetrameric complex (stoichiometry A2B2). Includes FGFR1 and FGFR2; FGF1, FGF2, FGF10. 5 CFs, only 2 are annotated in PDB as tetramer. Rest as heterodimer. dunbrack2.fccc.edu/ProtCAD/Result… 

We include deposited biological assemblies (from authors and PISA) in our clustering. Here are actin filament assemblies (A3, A4, A5, A8) combined. 27 CFs, 30 PDB entries, 6 UNPs. Filaments are challenging to differentiate from crystallization-induced chains of monomers. 

Hormone receptor homodimers and heterodimers (estrogen receptor, PPAR-gamma, RXR heterodimers, etc.). 79 CFs, 39 UNPs, 347 entries. Dimer is in PDB biological assemblies for only 226 of these entries (65%). Not sure why so many are missing. 

(Neur_chain)_(Neur_membrane) Pfam architecture. Pentamers of ligand-gated ion channels. 39 CFs, 21 UNPs, 144 PDB entries. 

GPCR Galpha complexes with bound antibodies in gray (most such complexes). 31 CFs (in two different clusters with V_set (nanobodies) and (V_set)_(V_set) (scFvs)). 

The U1 snRNP, including complex of 7 human proteins: SNRPB, SNRPD1, SNRPD2, SNRPD3, SNRPE, SNRPF and SNRPG complex. 18 crystal forms, 26 entries, 10 species. 19 homoheptamers (mostly Archaea). 7 heteroheptamers (S. cerevisiae and Ss. pombe; human). 

• • •
Missing some Tweet in this thread? You can try to
force a refresh







