SARS-CoV-2 lineages, Cryptic lineages, and a prediction of what the next dominant VOCs will be.
We sequence SARS-CoV-2 from wastewater using a different approach than most. We sequence smallish fragments of the genome so that we can tell if particular mutations are derived from the same virus. This way we are better able to determine if there is anything new circulating. 

About a year and a half ago we detected something from a St. Louis sewershed that didn't make sense. SARS-CoV-2 lineages whose sequence did not match anything that had been seen from patients. We called these 'cryptic lineage'. 

Wanting to know if this kind of thing was common, we started collaborating with @DrJDennehy to see if we could find anything similar in other places. NYC also had cryptic lineages. Not the same lineages, but lineages with similar characteristics.
nature.com/articles/s4146…
nature.com/articles/s4146…
By the end of 2021, we had identified 9 sewersheds (out of about 180 that were being routinely surveyed) that contained cryptic lineages, including a lineage from California with @RoseKantor.
medrxiv.org/content/10.110…
medrxiv.org/content/10.110…
My obsession with finding and characterizing these cryptic lineages accelerated in 2022. To date we have screened wastewater from around 700 sewersheds. In total we have found 24 putative lineages. I say putative because some of them we have only seen once.
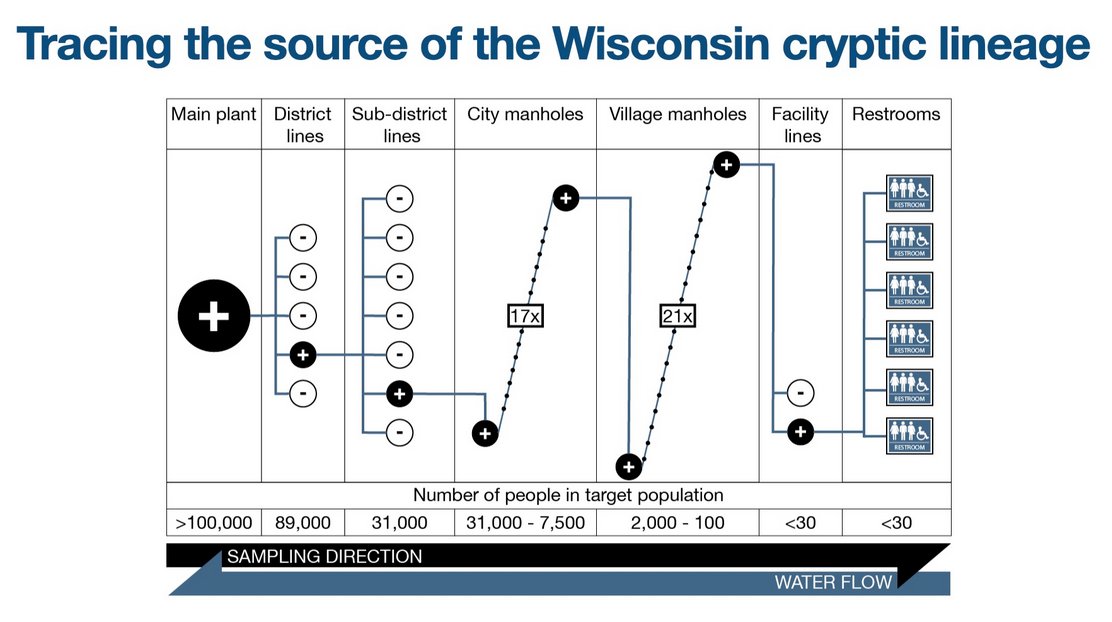
We still didn't know where these lineages were coming from. I was convinced of an animal reservoir. What changed my mind is when we tracked a lineage in Wisconsin with @dho all the way to a single set of bathrooms. There is no animal contribution. It is from a person. 
What we think is happening is that cryptic lineages are very long-term infections in people (often >1 year). Perhaps GI infections. The hosts are obviously mounting an immune response, but are not able to clear the infection.
Because there are no genetic bottlenecks from spreading from person-to-person, these viruses basically push the evolutionary fast-forward button.
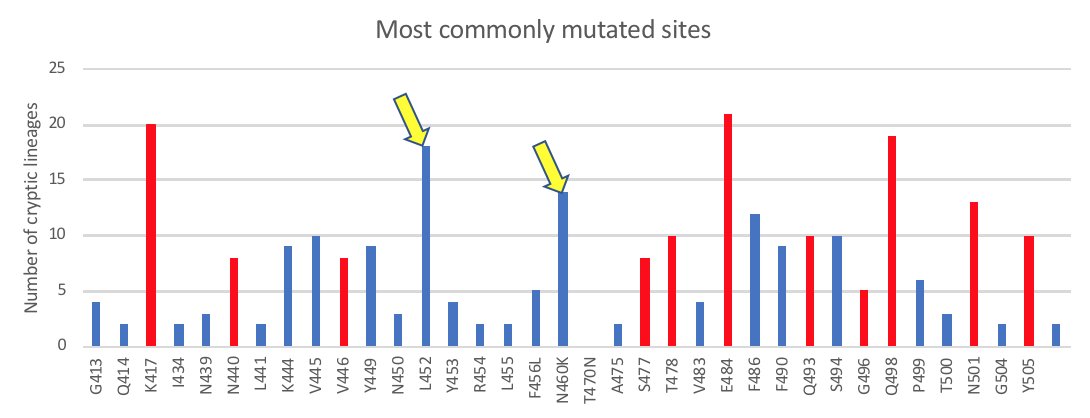
What can these lineages tell us about circulating viruses? These are the sites in the Spike RBD that are most commonly changed in the cryptic lineages. We had seen lineages with these changes long before they were seen in Omicron. 

When Omicron arrived (shown in red) it had mutations at many of the same sites. The main exceptions were L452 and N460, which were common in cryptic lineages, but were not in Omicron (BA.1). 
About six months later we started seeing Omicron lineages with mutations at L452. Highlighted in red are the changes in BA.4/5 (which has L452R), but there were also lineages with L452Q, L452M, and others. These lineages took over, but there were still no changes at N460. 
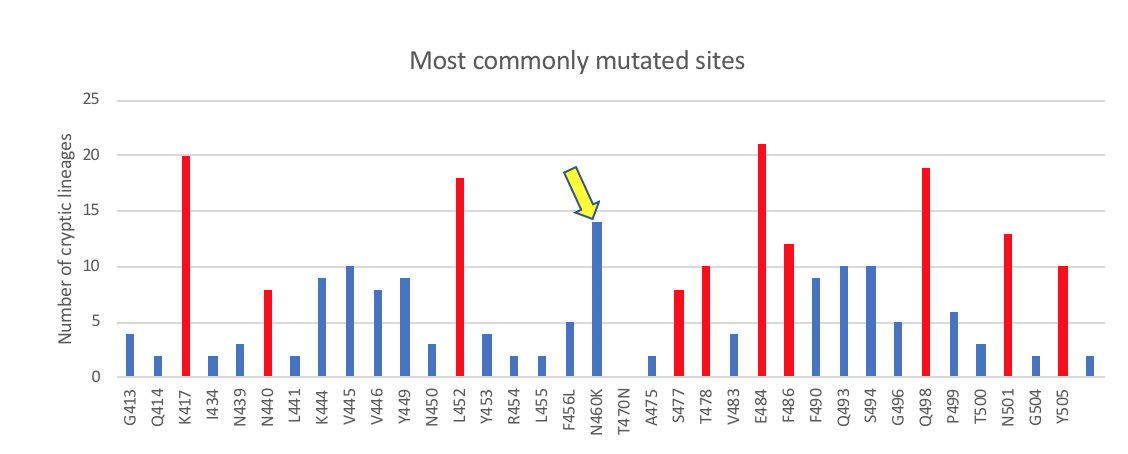
Finally, a few months ago N460K appeared in an Omicron lineage (BA.2.75). However, it was not combined with any of the L452 mutations. Nonetheless, BA.2.75 and its derivatives have continued to expand and slowly displace other lineages. 

A few days ago, a new lineage appeared in a MO wastewater sample. Omicron with L452R+N460K (and K444M). This appears to match a new lineage that was just designated as BU.1 a few days ago. Currently there are only 13 sequences in GISAID with this combination of mutations. 

However, I recently learned from @CorneliusRoemer that I was behind in my lineages. There is another completely independent lineage that arrived at almost exactly the same combination. This lineage is designated BQ.1.1. This group has L452R+K444T (rather than K444M).
In addition, BQ.1.1 has also picked up R346T (also in BA.4.6). We haven't focused as much on this region of spike, but most cryptic lineages that we have checked also have a mutation at this site. It seems to be pretty critical. BQ.1.1 is new, but it seems to be taking off.
There you have it. BQ and BU. I don't know if they will cause a spike in total cases, but barring something completely new appearing, I predict that they will be the dominant VOCs in the coming months.
Addendum. It seems a few other lineages have hit very similar 'jackpot' combinations very recently. In addition to BU and BQ:
BW.1 (BA.5.6 derivative)
BS.1 (BA.2.3 derivative)
BR.2 (BA.2.75 derivative)
other undesignated
They all have N460K, L452R + other changes.
BW.1 (BA.5.6 derivative)
BS.1 (BA.2.3 derivative)
BR.2 (BA.2.75 derivative)
other undesignated
They all have N460K, L452R + other changes.
Real pango experts, please correct me if I botch any of these, or have missed any. @CorneliusRoemer @PeacockFlu @siamosolocani @LongDesertTrain
• • •
Missing some Tweet in this thread? You can try to
force a refresh