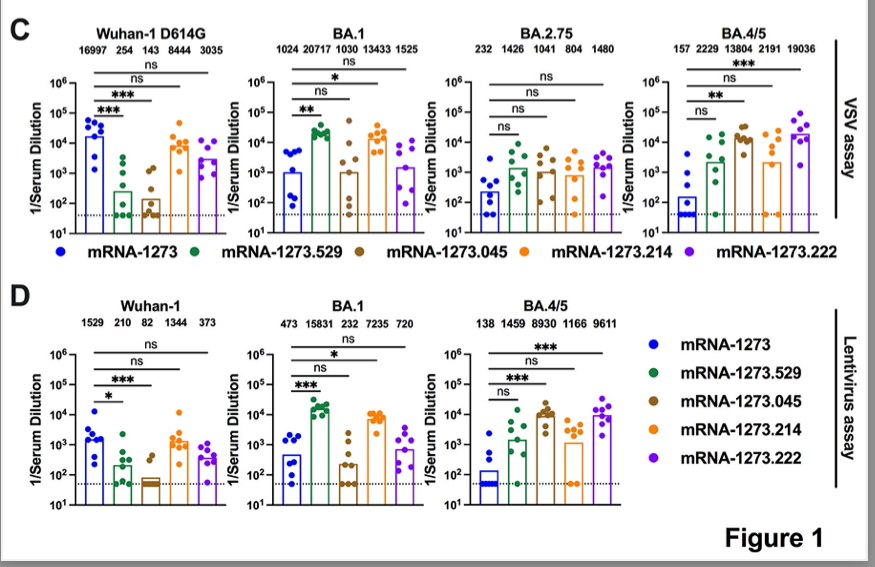
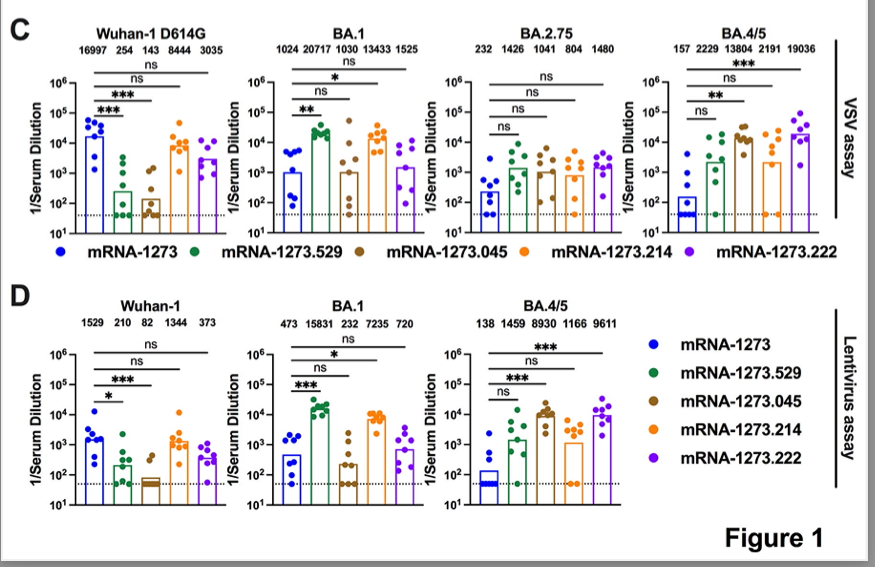
This study on response to Omicron after #SARSCoV2 vaccination provides some optimism re “original antigenic sin” biorxiv.org/content/10.110…
TLDR: although response to Omicron dominated by preexisting B-cells, antibodies from these cells better at 6 months than 1 month.
🧵 below.
TLDR: although response to Omicron dominated by preexisting B-cells, antibodies from these cells better at 6 months than 1 month.
🧵 below.
The study is by @chengzi_kaku & @LauraWa27423872, w contributions from others including @tylernstarr and myself.
It is highly complementary to recent studies by @yunlong_cao et al (
It is highly complementary to recent studies by @yunlong_cao et al (
https://twitter.com/yunlong_cao/status/1570922875590414336) and @TheBcellArtist (
https://twitter.com/TheBcellArtist/status/1573167965742239744).
It is now established that response to Omicron booster vax or breakthrough infection is dominated by boosting pre-existing B cells; see below.
(Studies differ on if it’s just dominated or overwhelmingly dominated, maybe due to differences including CoronaVac vs mRNA vax?)
(Studies differ on if it’s just dominated or overwhelmingly dominated, maybe due to differences including CoronaVac vs mRNA vax?)
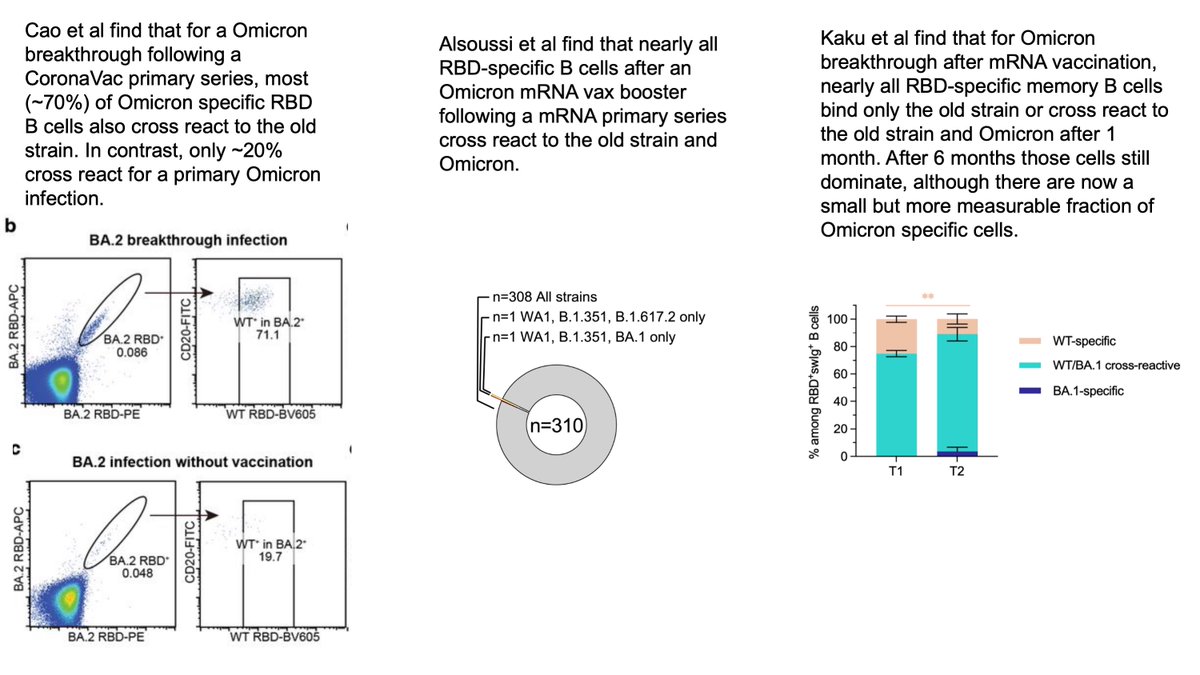
It’s not immediately obvious whether dominance of boosting cross-reactive B cells vs generating new ones is “good” or “bad” thing.
But the concerning observation is that resulting antibodies are often less (or even non) neutralizing to Omicron:
But the concerning observation is that resulting antibodies are often less (or even non) neutralizing to Omicron:
https://twitter.com/yunlong_cao/status/1570922944007901184
Importantly, this finding is shared across the other two studies: the cross-reactive antibodies elicited by Omicron boosting generally don’t neutralize Omicron as well as they neutralize the “old” viral strain: 
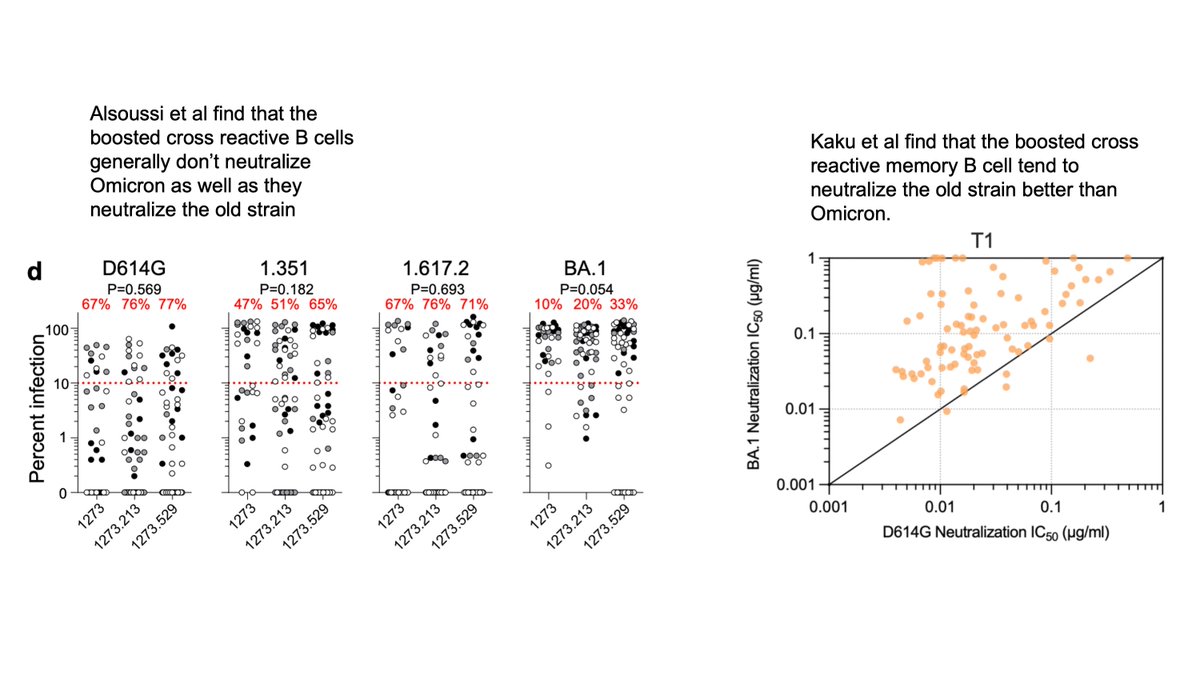
But unique aspect of Kaku et al study is to compare Omicron neutralization by antibodies from B cells at 1 versus 6 month after breakthrough infection (Cao et al only looked 1 month).
After 6 months, antibodies have affinity matured to be much more potent against Omicron!
After 6 months, antibodies have affinity matured to be much more potent against Omicron!
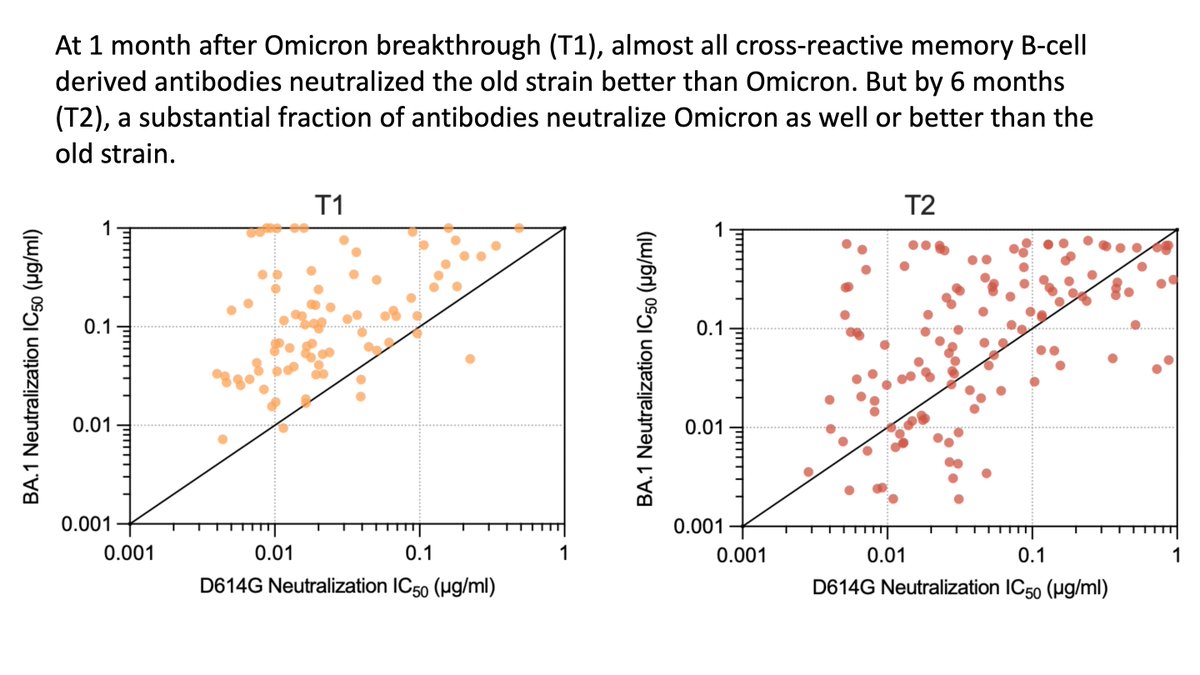
Improved neutralization of Omicron due to improved affinity for its RBD, which comes at slight cost to affinity for old RBD.
Shows affinity maturation favors mutations that “specialize” on Omicron vs old strain. But this is good, because we now care about Omicron, not old strain
Shows affinity maturation favors mutations that “specialize” on Omicron vs old strain. But this is good, because we now care about Omicron, not old strain
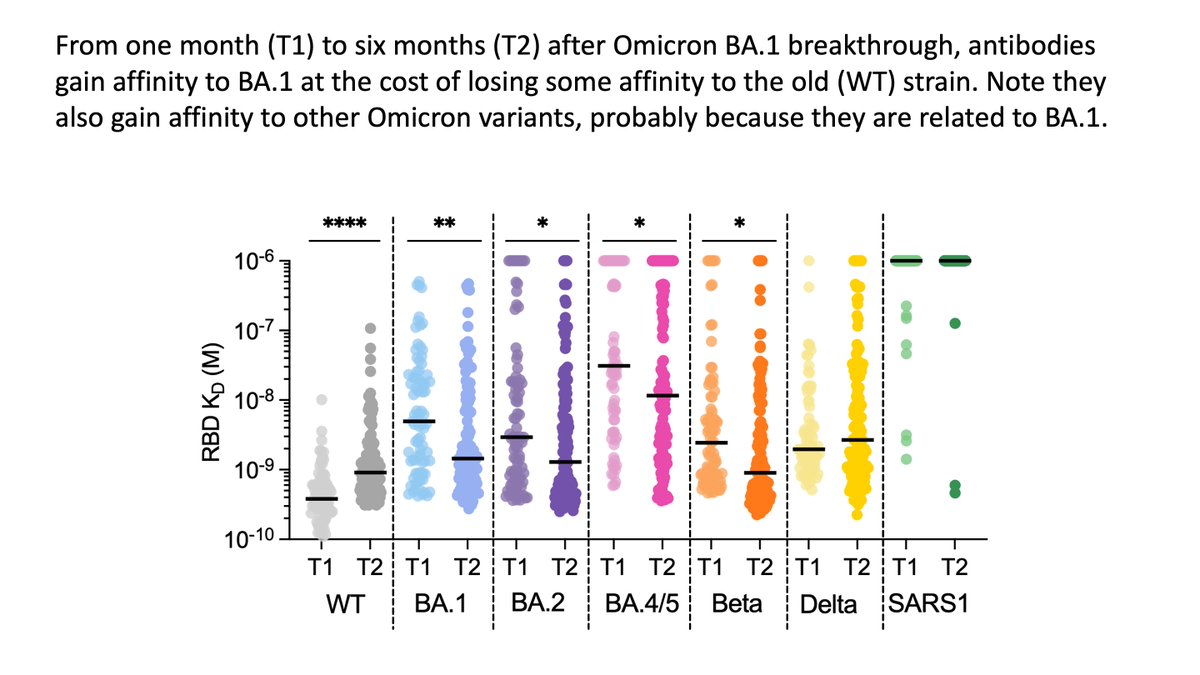
(Note that there is lots of good prior work, such as this study by @NussenzweigL et al (cell.com/immunity/fullt…), on long duration of affinity maturation to #SARSCoV2; what’s new in current study is showing it shifts neutralization towards Omicron after breakthrough.)
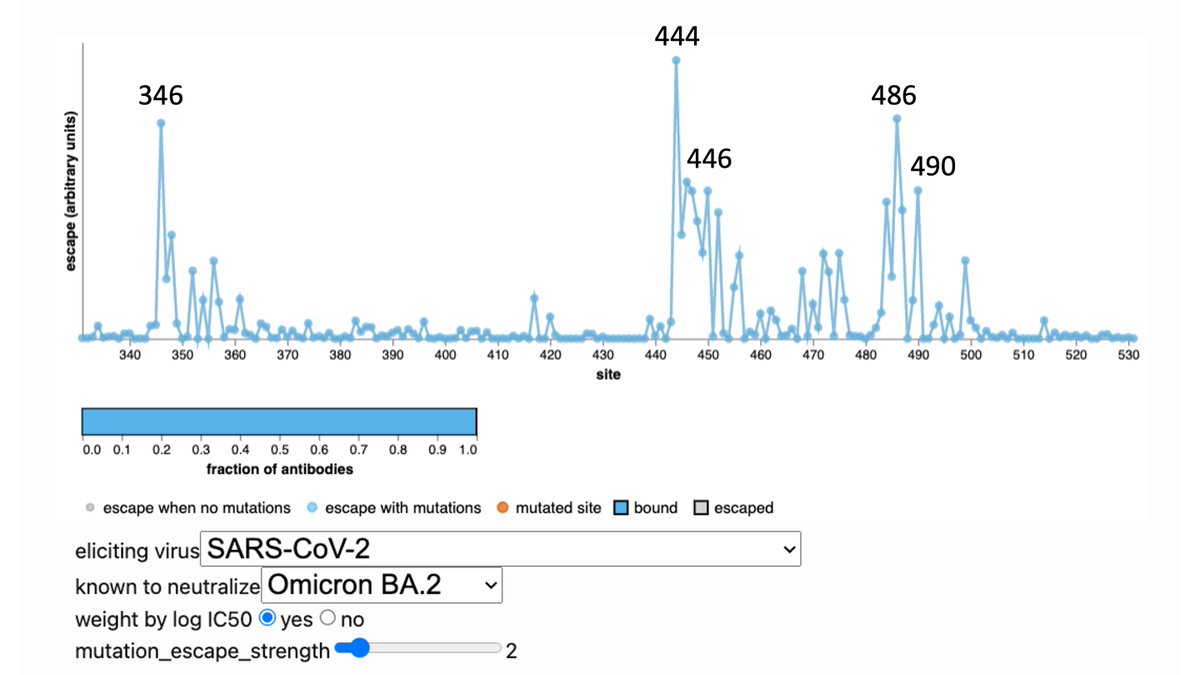
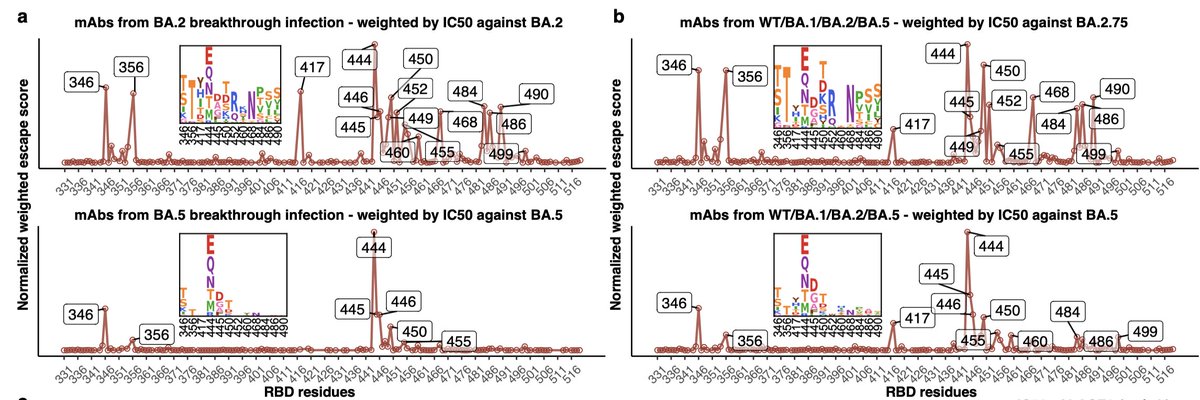
A few antibody classes dominate >50% of the Omicron breakthrough antibody response. Our contribution was deep mutational scanning on representative antibodies to map escape mutations. These include sites 346, 460, 486, 490, which are evolving in recent variants. 
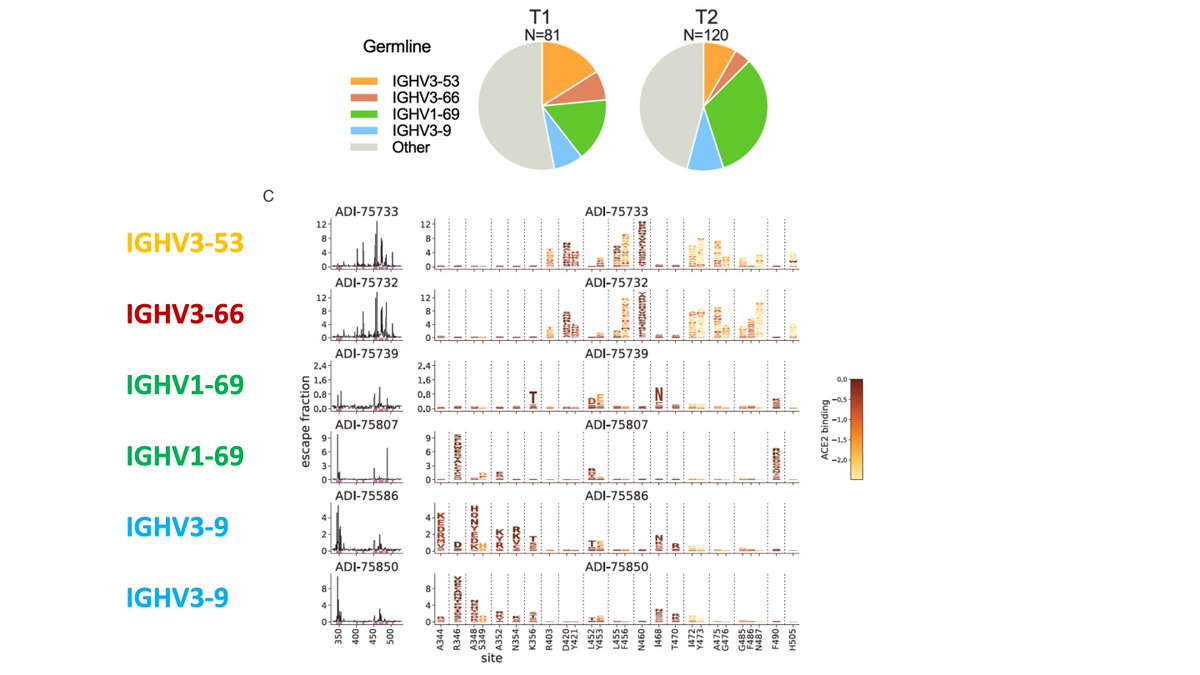
Our deep mutational scanning largely consistent w much larger study by Cao et al: antibodies that neutralize Omicron focus on a few sites that are now rapidly evolving:
(Aficionados: N460K, thought of as stability mutation, also escapes in our data.)
https://twitter.com/yunlong_cao/status/1570922968586551301
(Aficionados: N460K, thought of as stability mutation, also escapes in our data.)
However, it’s not clear that this unfortunate narrow neutralization focus on a few sites is new feature of Omicron-boosted antibodies.
Recall single E484K mutation caused a lot of escape from original “non-antigenic sin” antibodies to old strain:
Recall single E484K mutation caused a lot of escape from original “non-antigenic sin” antibodies to old strain:
https://twitter.com/jbloom_lab/status/1346442000472580098
Rather, it may be that a narrow neutralization focus that is susceptible to a handful of escape mutations is an unfortunate general feature of the antibody response to viruses with a high capacity for antigenic evolution:
https://twitter.com/jbloom_lab/status/1385664864765444098
Big picture: I think these studies suggest heterologous boosting, as from breakthrough or updated vaccine *is* helpful: affinity maturation towards Omicron & at least some elicitation of new B cells.
Seems to me boosting w updated strain is clearly preferable to not updating.
Seems to me boosting w updated strain is clearly preferable to not updating.
It’s possible that boosting with Omicron is less preferable than some magical world where we could turn back the immunological clock and give everyone primary immunity to the current circulating variant.
But that magical world doesn’t exist. At this point, most people have immune imprinting to #SARSCoV2 from vaccination, infection, or both. And virus will keep evolving, so long term everyone who is not a young child will be imprinted with an “old” strain for most of their lives.
Their imprinted immunity will get boosted at least every few years by a new strain through vaccination with updated booster (my preference, as I’d rather have sore arm than COVID-19), infection with the latest variant, or both.
This boosting will be on background of immunity from prior vaccinations/infections: but above studies show that boosting w new strains do offer beneficial affinity maturation and some new B cells.
Maybe (?) these benefits will compound over more Omicron exposures.
@victora_lab fate mapping study found that *two* Omicron exposures were necessary to start producing substantial levels of antibodies from “new” B cells.
@victora_lab fate mapping study found that *two* Omicron exposures were necessary to start producing substantial levels of antibodies from “new” B cells.
https://twitter.com/victora_lab/status/1564763238469877760
So while it’s possible updated boosters aren’t as good as magical world in which primary immunity is always to circulating variant, they seem better than real-world alternative of waiting for an infection to start updating immunity to new strain.
(But I do think it seems sensible to update the *primary* vaccination series being given to unvaccinated individuals, at this point mostly young children, to the new vaccine. This will start primary immunity as close as possible to where we’d like to be.)
• • •
Missing some Tweet in this thread? You can try to
force a refresh