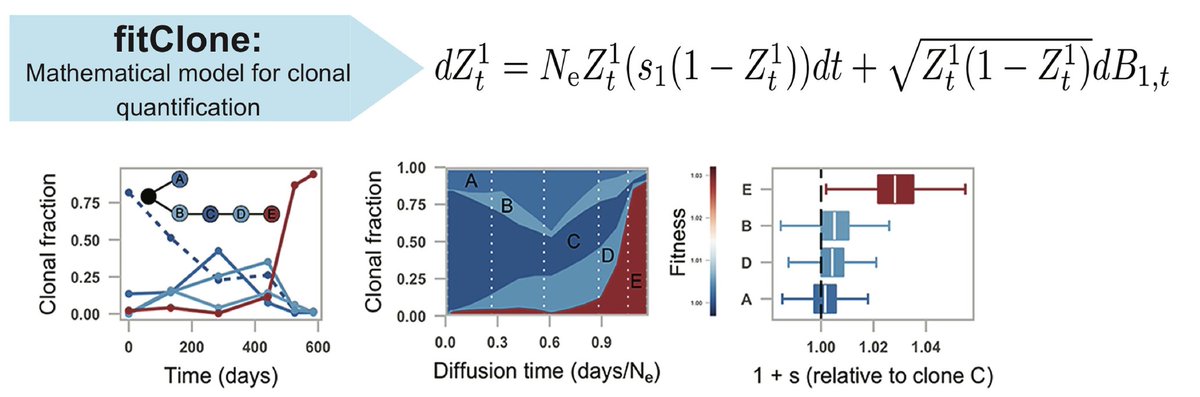
Out today in @Nature : nature.com/articles/s4158… - some steps toward understanding cell-to-cell genomic variation in cancer. Joint work with @sajraparicio lab (now 15yr collab!). Work performed by @tylerfunnell @marcjwills_ @oflanach and teams from both labs.
We studied how different background endogenous mutational processes impact individual cell genomes in cancer. Events in single cells represent to a first approximation the most recent alterations and so reflect the true diversity upon which selection can act.
We studied breast and ovarian cancers and identified three previously hidden states of cell-to-cell genomic variation that could influence phenotype and evolution. We generated >35k single cell genomes from genetically engineered cell lines and from human PDX models.
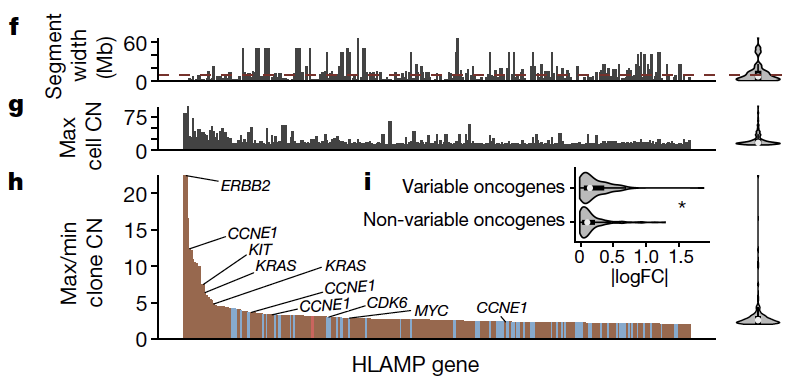
The first - we quantify the extent to which high level amplifications variable across clonal populations in the same tumor. This was observed at higher rates in foldback inversion bearing tumors relative to HRD and impacts major oncogenes. 

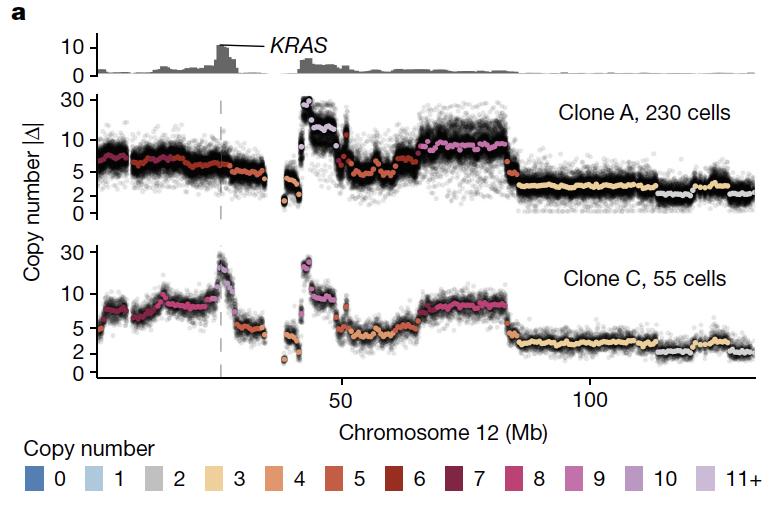
Second - we outline the degree of parallel copy number evolution whereby distinct maternal and paternal alleles impacting the same locus are altered in different cells in the same tumor. Parallel events on maternal and paternal alleles contributes to evolutionary diversity. 

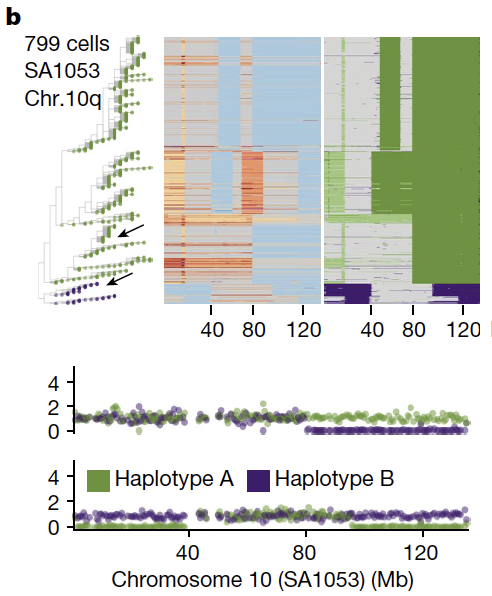
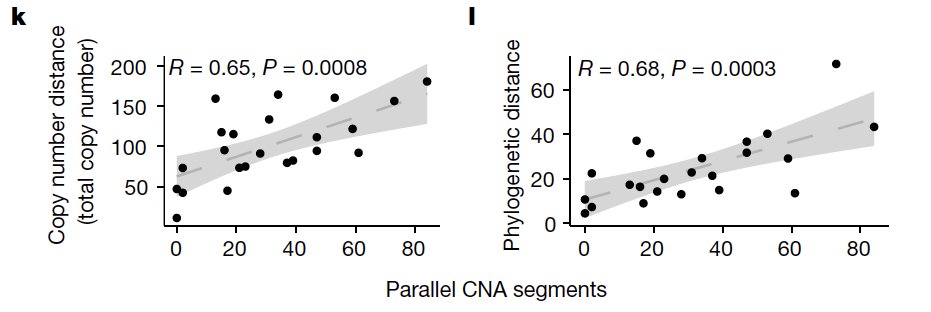
These findings are enabled by a computational method developed by @marcjwills_ called SIGNALS: github.com/shahcompbio/si…, building on previous advances from @benjraphael group.
Third- we uncover the degree to which ‘serriform structural variant (SSV)’ breakpoints whereby progressive changes in the breakpoints of structural variants from cell-to-cell indicate a clock-like SV mutational process. SSVs were also more prevalent in FBI tumors. 

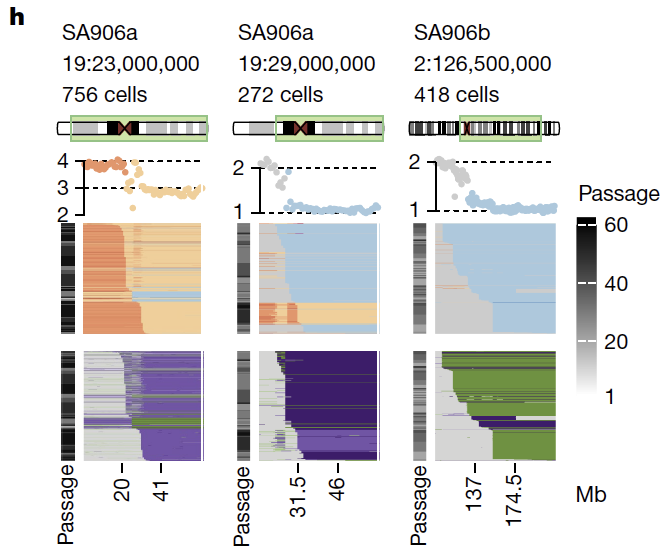
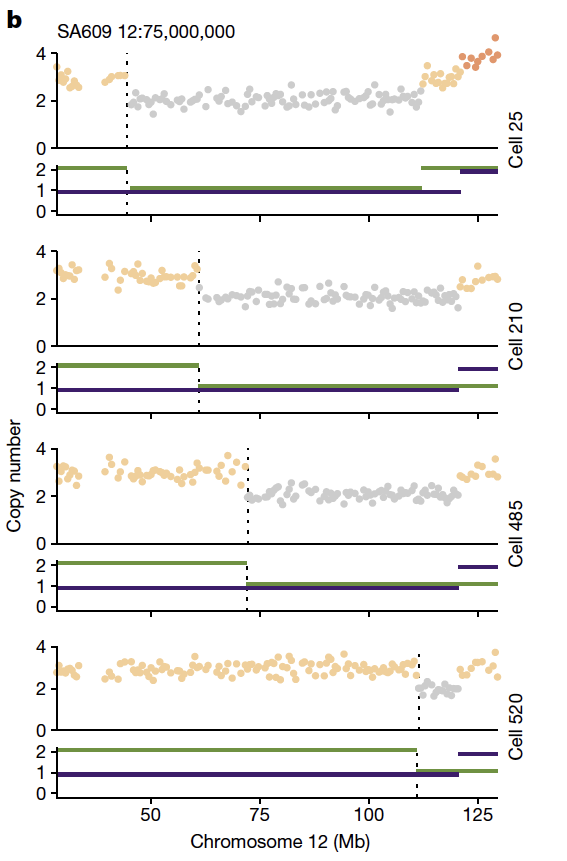
Overall the study illuminates a new dimension on how endogenous mutational processes impact 'evolvability' in cancer through profiling cell-to-cell genomic variation. How selection operates on this source of variation will be the subject of future studies on therapeutic response.
Huge thanks to @CancerGrand @Cycle4Survival @MSKCancerCenter @bccancerfdn for support and funding. Most importantly thank you to the patients and families for participating in this research.
• • •
Missing some Tweet in this thread? You can try to
force a refresh