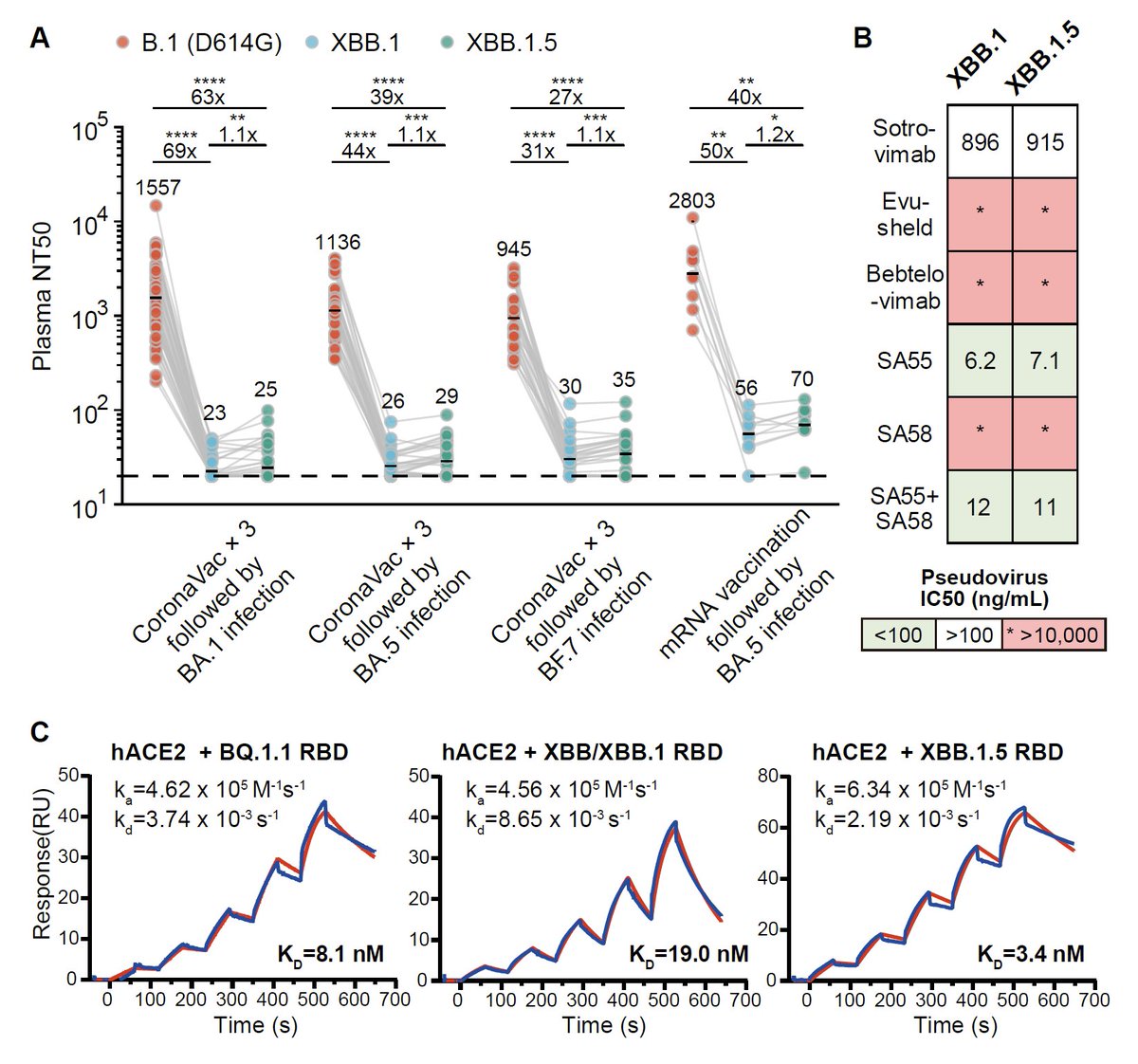
The superior growth advantage of XBB.1.5 has been well-documented by many colleagues @JPWeiland @LongDesertTrain @EricTopol. Here I'll add some experimental data:
1) XBB.1.5 is equally immune evasive as XBB.1, but
2) XBB.1.5 has a much higher hACE2 binding affinity. 1/
1) XBB.1.5 is equally immune evasive as XBB.1, but
2) XBB.1.5 has a much higher hACE2 binding affinity. 1/
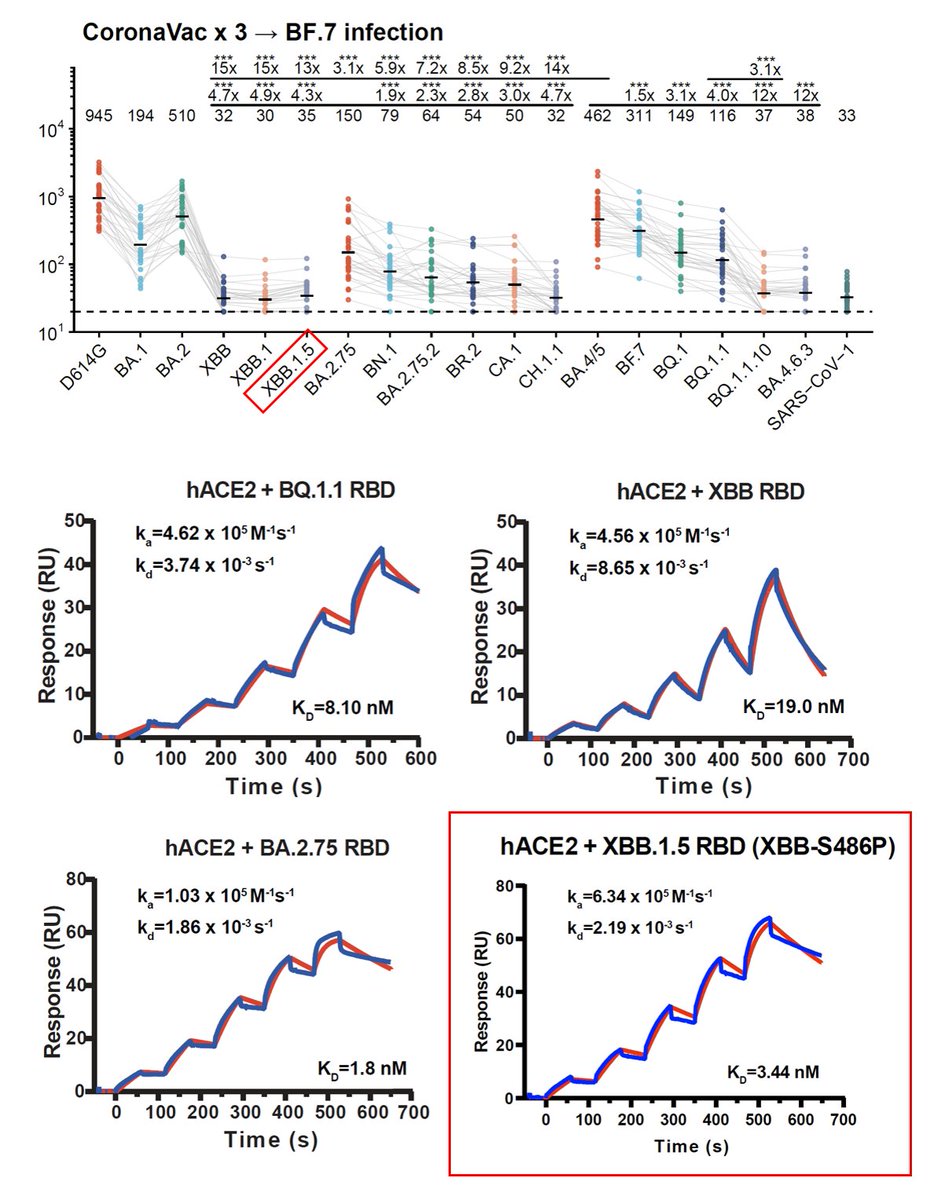
Notably, even BF.7 breakthrough infection doesn't induce high neutralization against XBB.1 and XBB.1.5. The S486P mutation only caused a slight reduction in immune evasion capability. mRNA breakthrough infection samples (n=9) here all received at least 2-dose mRNA vac. 2/ 

However, the S486P mutation greatly enhanced hACE2 binding, since 486S completely destroyed the local hydrophobic interaction while 486P retained it. 3/ 

The fact that XBB.1.5 showed a much superior growth advantage than XBB.1 suggests that hACE2 binding affinity does play a heavy role in SARS-CoV-2 spreading. XBB.1 truly suffered from low-hACE2 binding, despite XBB.1’s highest immune evasion capability. 4/
https://twitter.com/JPWeiland/status/1607835958388432896
Another important observation is that XBB.1.5's hACE2 binding affinity is almost comparable to that of BA.2.75, which may enable XBB.1.5 to gain more mutations, similar to what BA.2.75 had. It's just XBB.1.5 haven't felt much immune pressure yet. 5/
Detailed information for the experiments described here can be found in the newly uploaded preprint:
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
• • •
Missing some Tweet in this thread? You can try to
force a refresh