
Biochemistry & Immunology | Host Immune Response, Antibody Drug & Vaccine Design | PhD @Harvard | Assistant Professor at Peking University BIOPIC
18 subscribers
How to get URL link on X (Twitter) App


 Q493E of KP.3 may have an epistasis effect with the F456L mutation, resulting in the increase of ACE2 binding affinity. This is important since strong ACE2 binding would allow KP.3 to easily accumulate highly immune evasive RBD mutations, such as A475V.
Q493E of KP.3 may have an epistasis effect with the F456L mutation, resulting in the increase of ACE2 binding affinity. This is important since strong ACE2 binding would allow KP.3 to easily accumulate highly immune evasive RBD mutations, such as A475V. 

 Since JN.1 lineages have replaced XBB lineages and JN.1 subvariants are continuously gaining immune-evasive mutations, such as R346T, F456L, R346T+F456L (FLiRT), and F456L+Q493E (KP.3), it's time to evaluate whether we need to switch SARS-CoV-2 vaccine antigen to JN.1.
Since JN.1 lineages have replaced XBB lineages and JN.1 subvariants are continuously gaining immune-evasive mutations, such as R346T, F456L, R346T+F456L (FLiRT), and F456L+Q493E (KP.3), it's time to evaluate whether we need to switch SARS-CoV-2 vaccine antigen to JN.1. 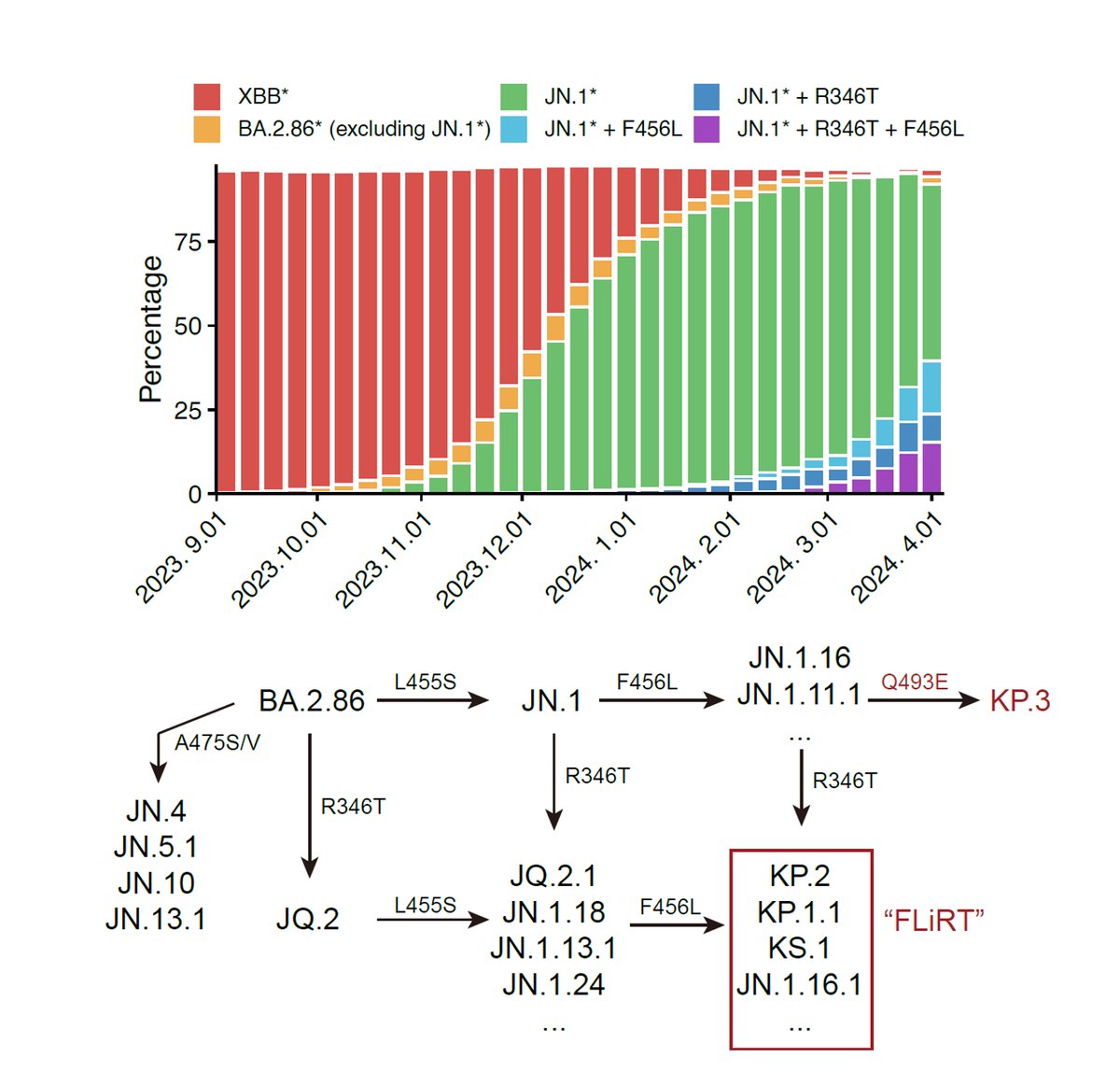


https://twitter.com/yunlong_cao/status/1714694253530804588

 Detailed descriptions of this paper can be found in our previous tweets published half a year ago:
Detailed descriptions of this paper can be found in our previous tweets published half a year ago:https://twitter.com/yunlong_cao/status/1653804041821093889

 BA.2.86's RBD showed a pretty high hACE2 binding affinity measured by SPR, higher than that of XBB.1.5 and EG.5 and is even comparable to "FLip" variants like HK.3. BA.2.86's V483del indeed decreases ACE2 binding, but R403K is just too powerful and makes up for the loss. 2/n
BA.2.86's RBD showed a pretty high hACE2 binding affinity measured by SPR, higher than that of XBB.1.5 and EG.5 and is even comparable to "FLip" variants like HK.3. BA.2.86's V483del indeed decreases ACE2 binding, but R403K is just too powerful and makes up for the loss. 2/n 

 By using pseudovirus neutralization assay and antigenic cartography (based on mRNA immunized mouse serum), we found that BA.2.86 is antigenically distinct from WT, BA.2, BA.5, and XBB.1.5. This means that XBB-induced antibodies cannot well recognize and neutralize BA.2.86. (2/n)
By using pseudovirus neutralization assay and antigenic cartography (based on mRNA immunized mouse serum), we found that BA.2.86 is antigenically distinct from WT, BA.2, BA.5, and XBB.1.5. This means that XBB-induced antibodies cannot well recognize and neutralize BA.2.86. (2/n) 

 The L455F+F456L RBD mutation combo is a very smart move by the virus (it's actually an LF->FL shift). Note that both individual L455F or F456L actually lose ACE2 binding, but together, the LF->FL shift somehow strengthened ACE2 interaction while destroying most antibody binding.
The L455F+F456L RBD mutation combo is a very smart move by the virus (it's actually an LF->FL shift). Note that both individual L455F or F456L actually lose ACE2 binding, but together, the LF->FL shift somehow strengthened ACE2 interaction while destroying most antibody binding. 


 Like the results by Kei @SystemsVirology, we found XBB.1.16 and XBB.1.5 have comparable immune evasion capabilities in the serum tested. The ACE2 binding affinity of XBB.1.16 and XBB.1.5 is also similar. In contrast, F456L brings additional immune evasion but lowers ACE2 binding.
Like the results by Kei @SystemsVirology, we found XBB.1.16 and XBB.1.5 have comparable immune evasion capabilities in the serum tested. The ACE2 binding affinity of XBB.1.16 and XBB.1.5 is also similar. In contrast, F456L brings additional immune evasion but lowers ACE2 binding. 
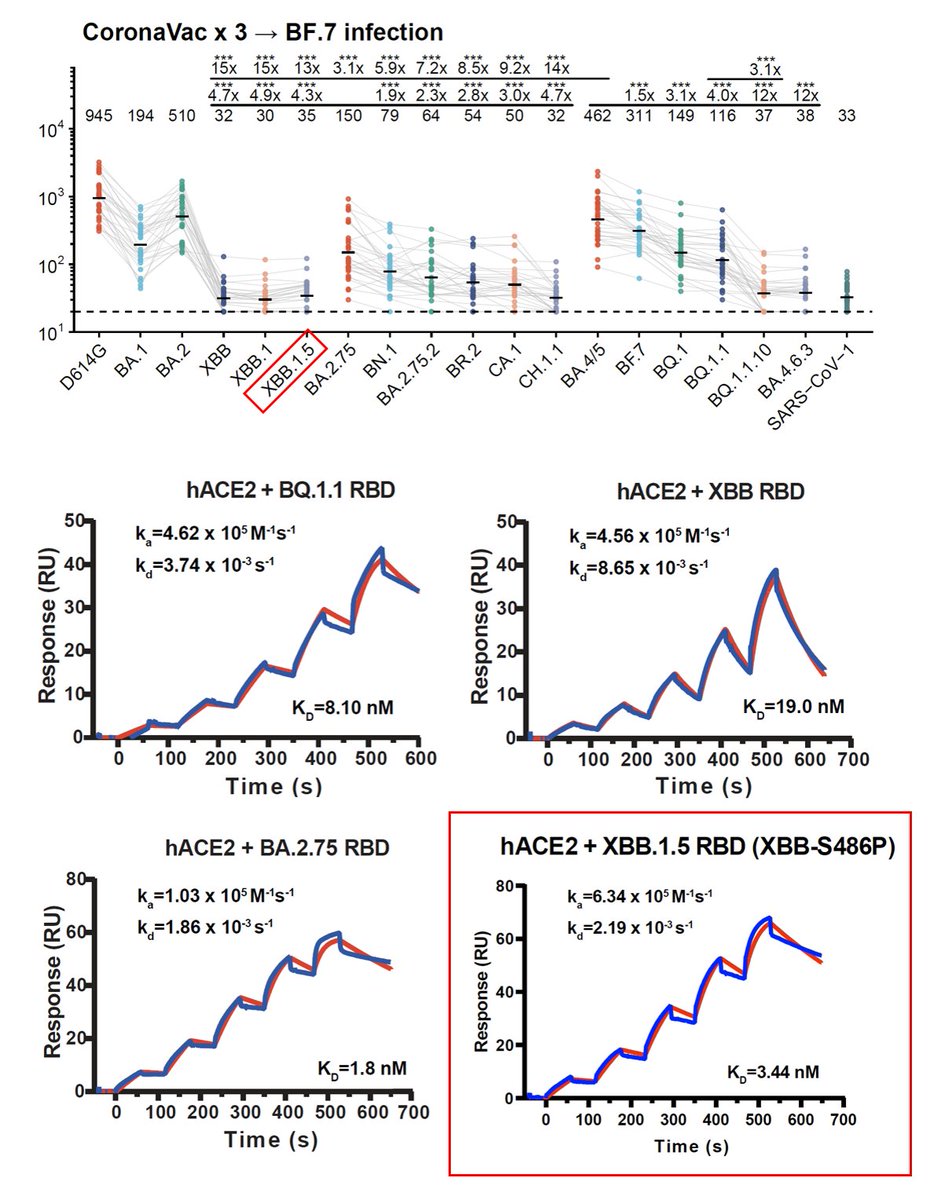
 Notably, even BF.7 breakthrough infection doesn't induce high neutralization against XBB.1 and XBB.1.5. The S486P mutation only caused a slight reduction in immune evasion capability. mRNA breakthrough infection samples (n=9) here all received at least 2-dose mRNA vac. 2/
Notably, even BF.7 breakthrough infection doesn't induce high neutralization against XBB.1 and XBB.1.5. The S486P mutation only caused a slight reduction in immune evasion capability. mRNA breakthrough infection samples (n=9) here all received at least 2-dose mRNA vac. 2/ 

 Like @LongDesertTrain @JosetteSchoenma @CorneliusRoemer have mentioned, recently there has been a rapid increase of Y144del proportion in the BQ.1* lineages. This NTD deletion is observed in many worrisome BA.5 sublineages such as BQ.1.1.10, BQ.1.18, as well as BA.4.6.3.
Like @LongDesertTrain @JosetteSchoenma @CorneliusRoemer have mentioned, recently there has been a rapid increase of Y144del proportion in the BQ.1* lineages. This NTD deletion is observed in many worrisome BA.5 sublineages such as BQ.1.1.10, BQ.1.18, as well as BA.4.6.3. 
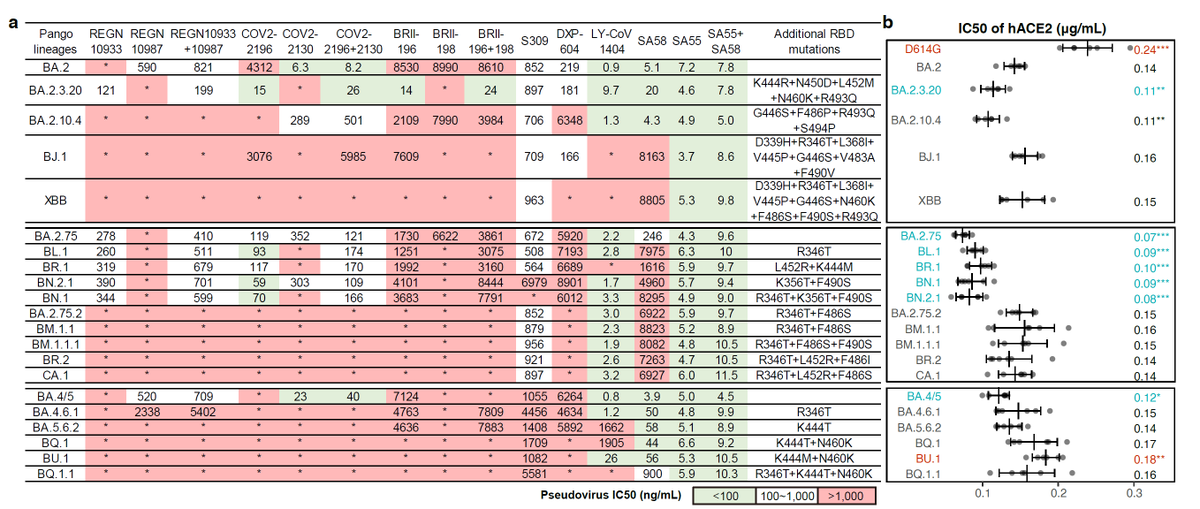


https://twitter.com/yunlong_cao/status/1570922875590414336BA.2.75.2 is slightly more evasive than BQ.1.1 against plasma from BA.2/BA.5 breakthrough infections. Its due to the enriched NTD-NAbs elicited by BA.2/BA.5 infections, which BQ.1.1 can't escape. Note that these varaints are approaching SARS-CoV-1 level escaping capability. (2/4)



