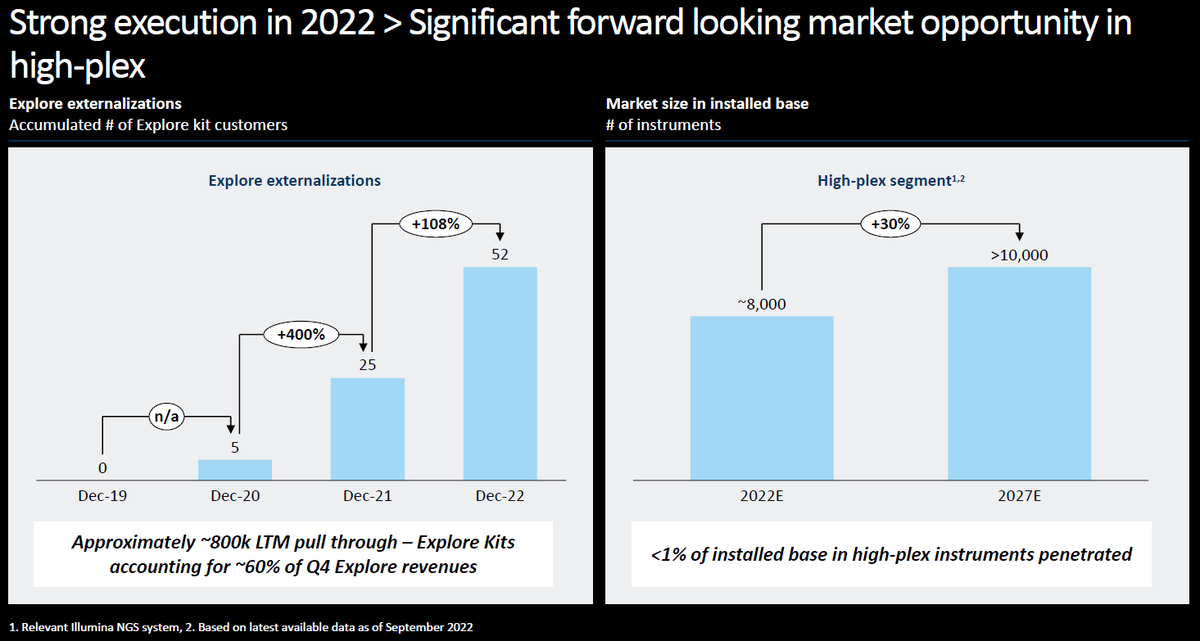
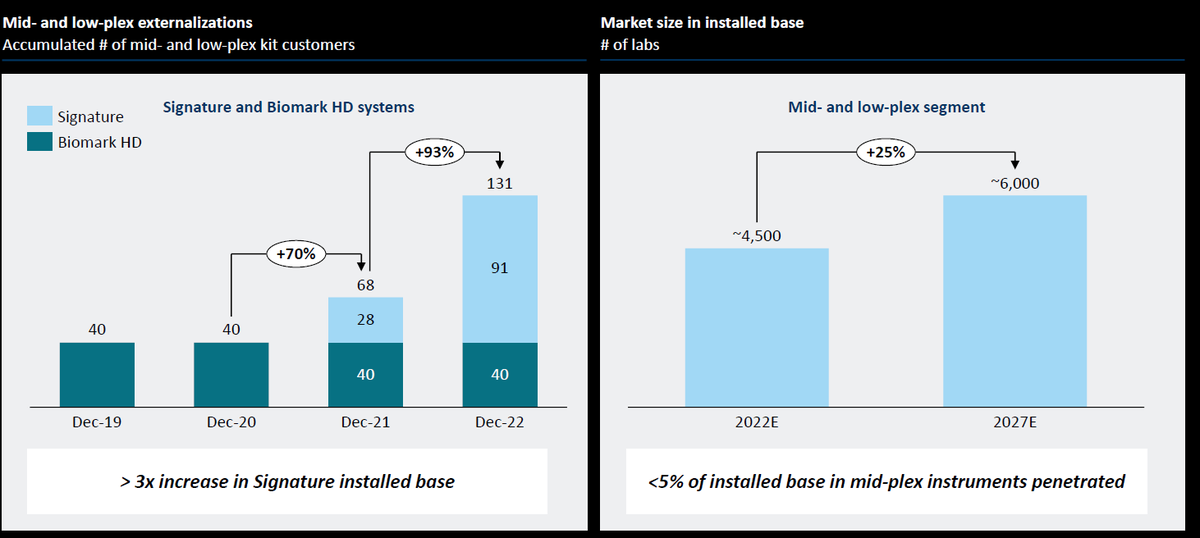
In #JPM2023, $NAUT Nautilus Bio didn't make their slides available, but they have a slide deck from an investor meeting in December 2022. They intend to launch their Proteome Analysis Platform in Mid-2024. 

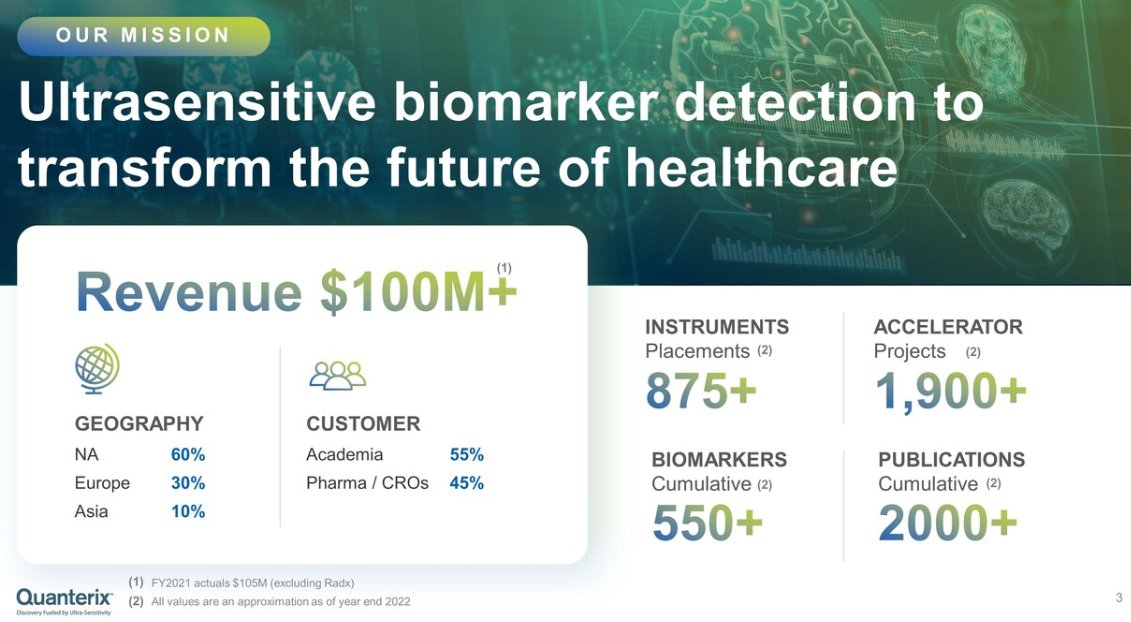
They see a market opportunity of $25B, where 50% would be BioPharma customers, and 20% Academic and Research. 

One of the biggest piece of news is that $NAUT Nautilus Bio recently partnered with Abcam to enhance their affinity reagent development program. 

They still have both aptamers and antibodies described as their affinity reagents, but Abcam would contribute and give weight to the antibody-based mode of generating more data points from proteome samples. 
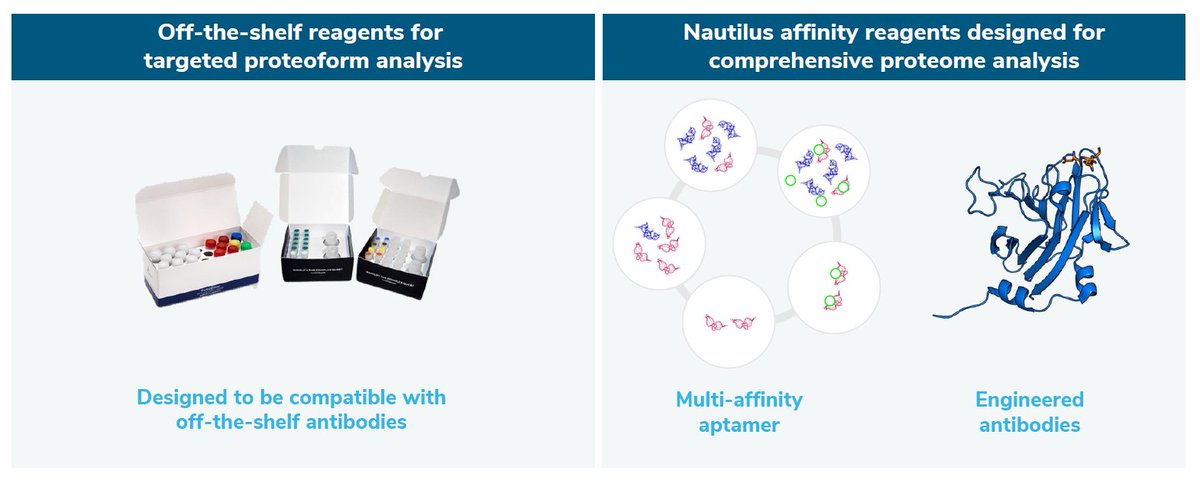
Their sequential affinity reagent hybridization and imaging is not too dissimilar from other approaches in NGPS, although the devil is in the details. 

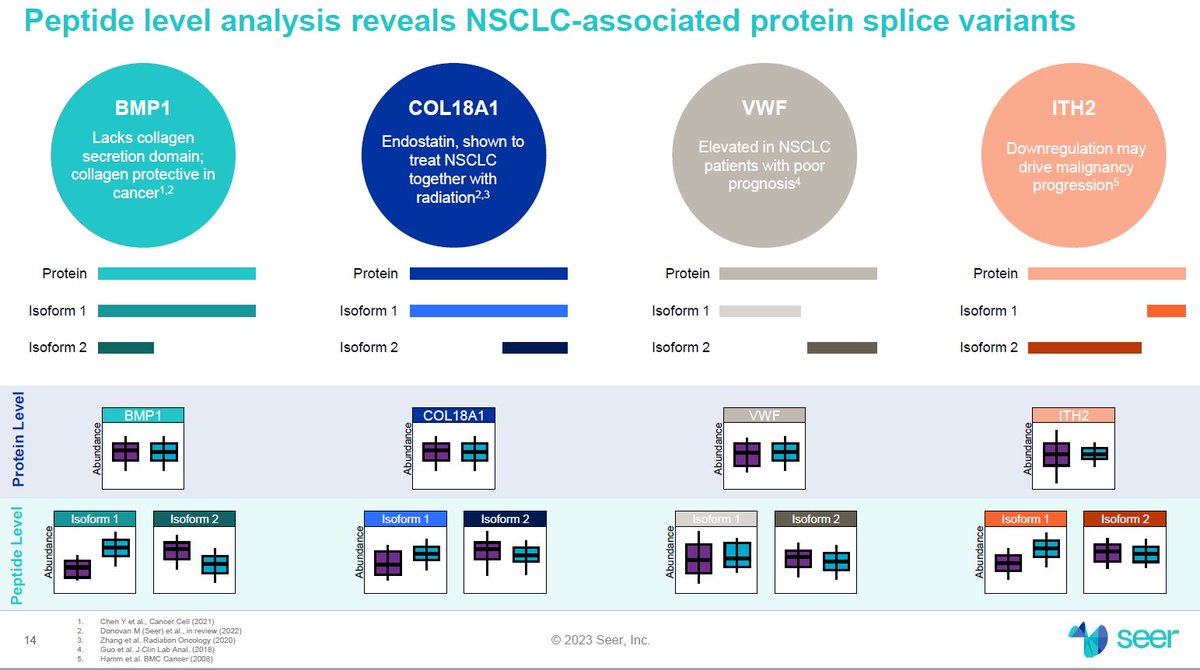
They can also do what they call Proteoform Mapping, which is a more targetted approach towards protein and PTM identification 

More on NGPS at bit.ly/ngps-slides
• • •
Missing some Tweet in this thread? You can try to
force a refresh