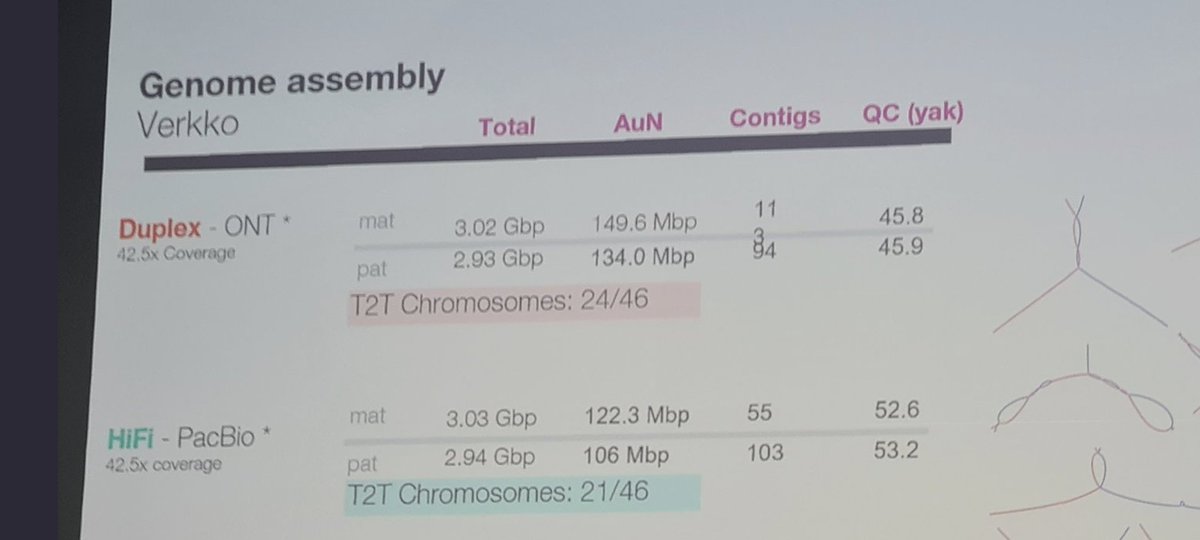
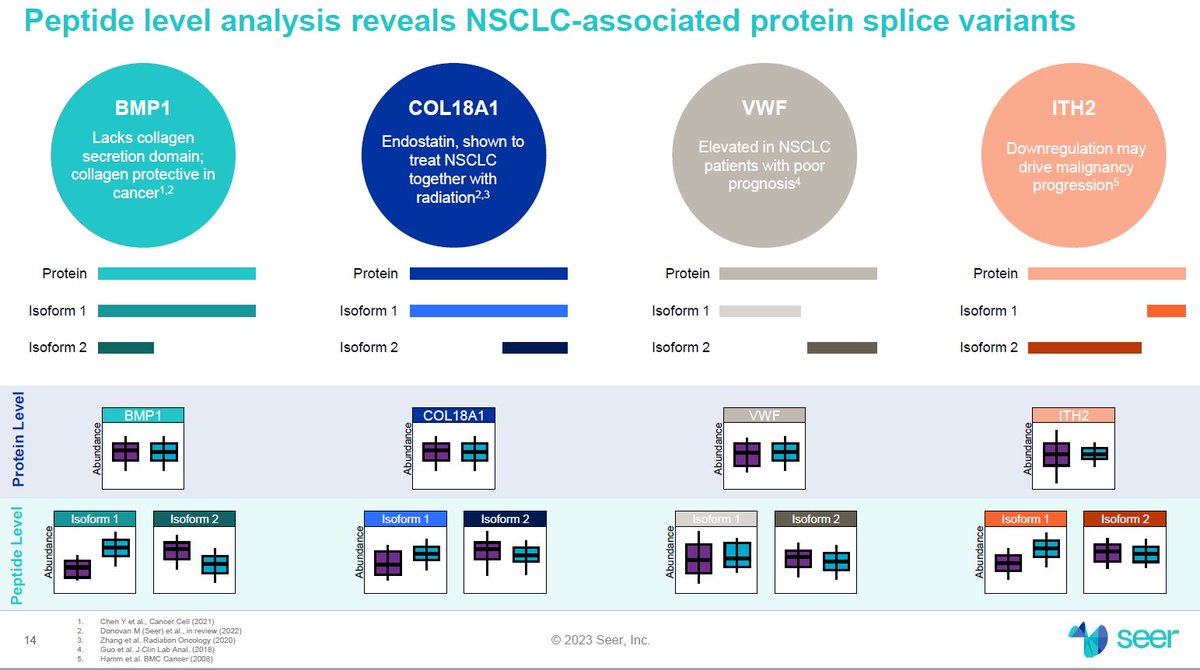
#AGBT23 More details on the $ILMN Illumina Infinity reads technology from Alex Aravanis (post from LinkedIn). 

Also first data shared on XLEAP-SBS chemistry for the NextSeq 1000/2000 instruments. Not sure why would anyone buy one of these instruments given the alternatives from @ElemBio and @CompleteGenomic 's G400. 

First shipment on NovaSeq X: if Illumina can beat the competition at anything in the next few months, it'll be on manufacturing and deployment of the NovaSeq X. 

Link to post: linkedin.com/posts/alex-ara…
• • •
Missing some Tweet in this thread? You can try to
force a refresh