The world of Next-Generation Sequencing (NGS) post-#AGBT23:
1) Some will say that $ILMN Illumina is sleep-walking into a cliff: the company has been dominating the field with 80-90%+ of the market-share, but they are unable to retain their technological advantage to competitors:
1) Some will say that $ILMN Illumina is sleep-walking into a cliff: the company has been dominating the field with 80-90%+ of the market-share, but they are unable to retain their technological advantage to competitors:
- Illumina doesn't have the most affordable $/Gb platform anymore, currently at $3.2/Gb, and $2/Gb in H2 2023, but others are already at $2/Gb, $1.5/Gb and will be at $1/Gb in Q3 2023.
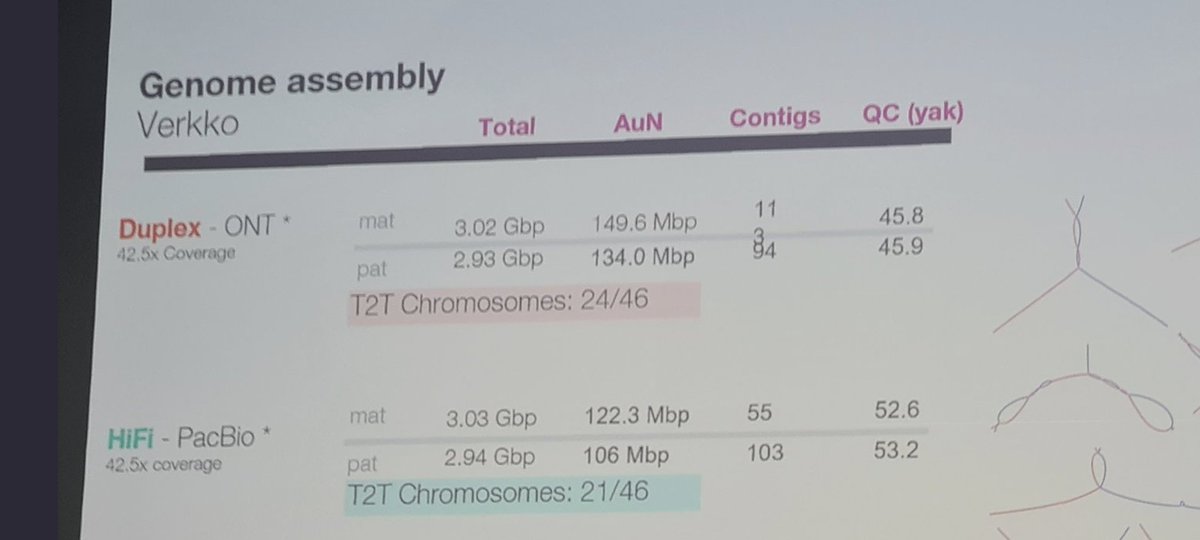
- Illumina doesn't have the longest read technology, or anything near the competition in terms of read length times $/Gb. Oxford @nanopore is unmatched with their ultra-long read technology and is nearing $10/Gb Q30+ performance, as is PacBio on 15-10Kb reads.
- Illumina doesn't have the most affordable sequencer in the low-throughput range, doesn't have the cheapest $/Gb in the mid-throughput range, or the cheapest high-throughput $/Gb or instrument.
- Illumina doesn't have their own technology for low-input or direct epigenetic 5mC base calling (PacBio does) or direct 5mC+5hmC calling (Oxford @nanopore does). This is important for cfDNA in Liquid Biopsy.
- Illumina won't be able to keep @GrailBio under the fold, unless risking heavy fines from the EU soon, and possibly the US regulators later. Their vertical in Liquid Biopsy for Cancer Screening is under threat from $EXAS Exact Sciences, $GH Guardant Health and others.
2) We have well and truly entered the $200/genome era, with several companies already pricing at that point, and such era will be short lived by the $100/genome era that we are going to enter later in the year. If @UltimaGenomics releases the U100 in 2023, there will be several
offerings in the $100/genome range for short-reads. This new pricing will make the market for NGS grow, especially the applied market, with biotech and clinical diagnostics growing significantly.
If this burst of new entrants and assays had happened 2 or 3 years ago, I would worry about an NGS bubble forming. But we have been in a depressed NASDAQ for the last 1.5-2 years, so all the companies with an interest in the field will have to have a sharpened business plan and
laser-focussed determination in applying NGS in a useful way. Those old enough to remember the dot-com bubble will also remember who survived the bubble bursting. The survivors of the dot-com bubble continued to apply similar technologies to similar markets, but the costs for
compute, storage and especially networking continued to drop post- dot-com bubble and the survivors, the Google's of the world, benefited from it. Will be see a renaissance of applied NGS post-#AGBT23?
3) Even if some consider the NGS landscape as being overcrowded, we still have large corporations with NGS aspirations that could make strategic acquisitions during this and next year. Some will criticize the new-comers for not having a big enough sales footprint, but that only
makes these M&A moves by big players more obvious, and they can rival Illumina in salesforce, in some cases outsizing them. One of the reasons the Thermo Fisher Ion Torrent technology is still around in decent numbers is their large sales presence.
4) The *short-reads* era of $100 genomes will only emphasize how important is to have a good reference genome to start with. Here "reference genome" will become "personal reference genome" on top of which personal annual liquid biopsy NGS check-outs can become a reality.
And although human health has always been the first to benefit from cheaper and cheaper NGS, AgBio is bound to benefit tremendously from this new era. Although we have accumulated dozens if not hundreds of reference genomes in AgBio, this is a field where Oxford @nanopore
technologies can make an even bigger difference than in any other field so far. Large plant genomes, with whole-genome duplication events and intricated epigenomic landscapes, have been underserved by a "too little, too short" NGS proposition from short-reads so far.
One of the biggest differences between plants and animals, think a field of wheat versus a gnu, is that plants can't run away from where they grow, so they benefit from having several copies of the similar gene families, also known as gene paralogues. When the rain patterns or
the temperature or soil acidity change, plants benefit from switching on and off certain copies of these gene paralogues, via epigenetic marks such as 5mC, and thus "adapt in place" to their new reality. In contrast, animals such as a gnu, have proportionally fewer copies of a
set of gene families, but can use alternative splicing to use the basic set of 20-25,000 genes to specialise in different cell types, tissues and organs. That includes the bones, nerves and muscles needed to "walk away" from an unfriendly environment, and find greener pastures.
So the larger more repetitive plant genomes will benefit from cheap ultralong @nanopore reads, capable of reading epigenetic marks natively, and do in highly deployable setups with the MinION and PromethION P2 Solo instruments.
(end of the post-#AGBT23 thread for now)
• • •
Missing some Tweet in this thread? You can try to
force a refresh