
The First Sequencing of the Bivalent vaccines is now available for public consumption.
WARNING- There are contaminating Expression vectors in the vaccines that contain antibiotic resistance genes.
This might explain the prolonged expression of spike protein in many studies
WARNING- There are contaminating Expression vectors in the vaccines that contain antibiotic resistance genes.
This might explain the prolonged expression of spike protein in many studies
This sequencing cost $7 per sample.
I wonder if these Pharma's can afford this for every lot?
Nah.. Why would they?
Your government gave them a liability shield and even paid them billions for the products they then mandated.
anandamide.substack.com/p/curious-kitt…
I wonder if these Pharma's can afford this for every lot?
Nah.. Why would they?
Your government gave them a liability shield and even paid them billions for the products they then mandated.
anandamide.substack.com/p/curious-kitt…
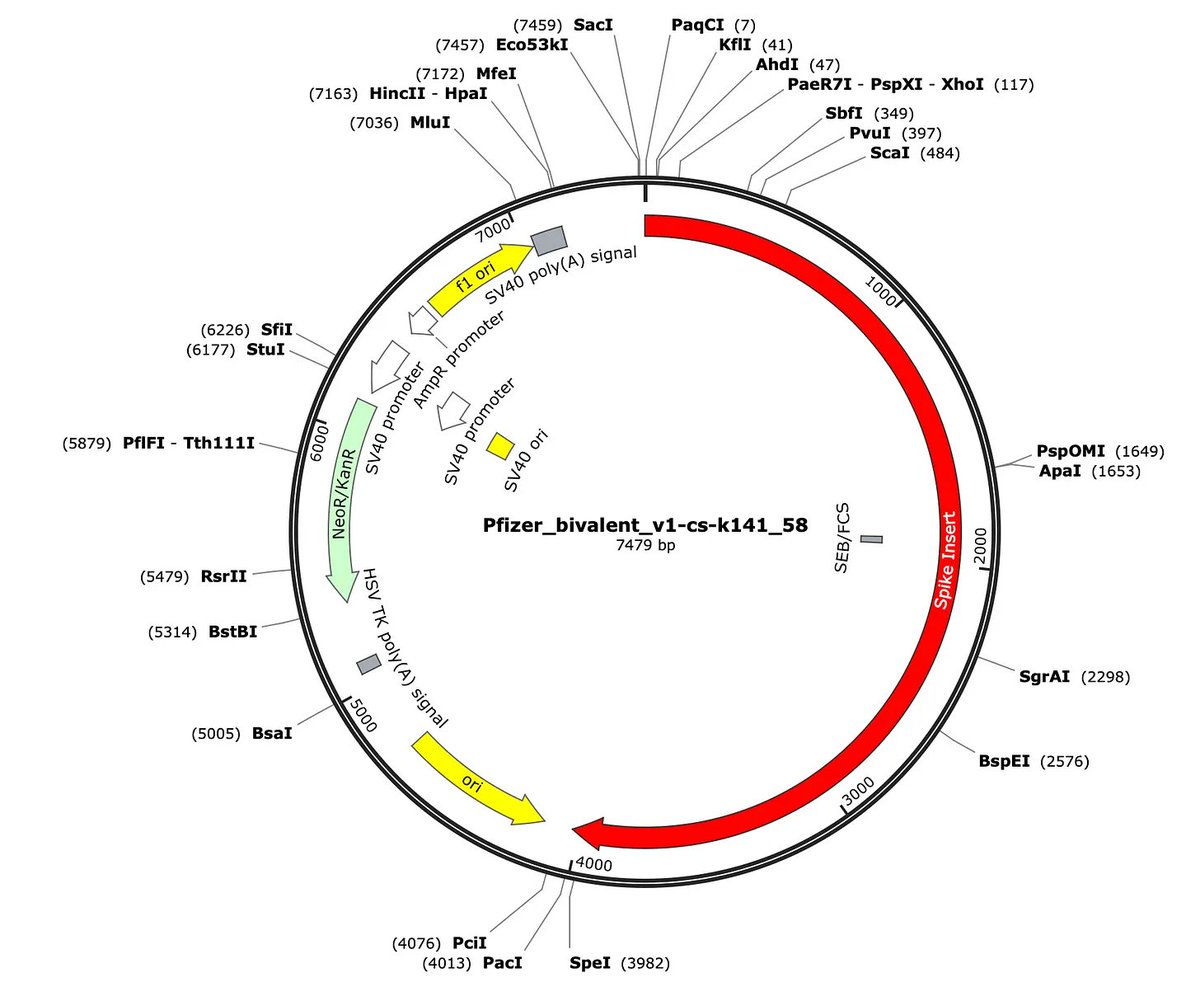
Among other treats in these expression vectors are the SV40 mammalian promoter and a high copy origin of replication (pUC) for bacterial amplification of these plasmids.
The Biodistribution studies covered by @Jikkyleaks show LNPs getting into the intestines.
The Biodistribution studies covered by @Jikkyleaks show LNPs getting into the intestines.
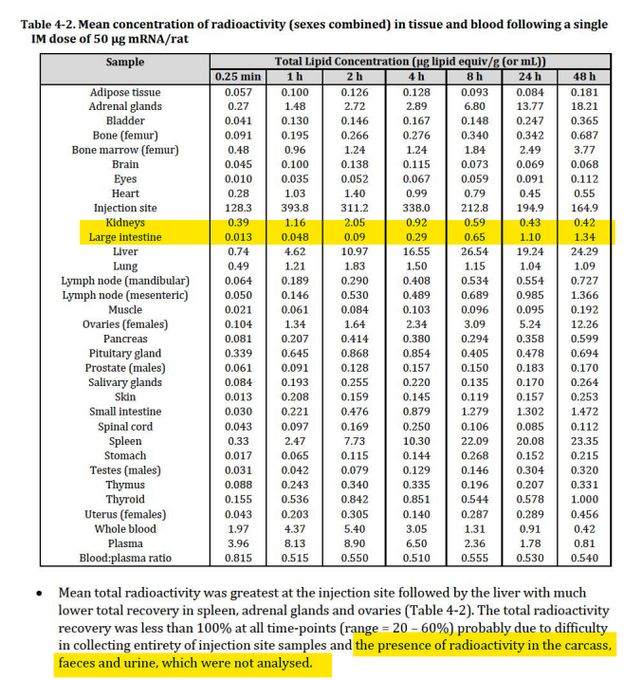
Its estimated that each shot contains billions of these plasmids and if they get into the gut microbiome, they can replicate to 50-300 copies per cell.
If any vaccinated patient is then placed on Neomycin or Kanamycin, these will be the only bacteria that survive
If any vaccinated patient is then placed on Neomycin or Kanamycin, these will be the only bacteria that survive
As these plasmid encode Kan and Neo resistance. Its not known if these contaminating plasmid can also express spike protein but some of the vectors have T7 tails so we cant rule that out.
What happens if we inject billions of Antibiotic resistance genes into billions of people?
What happens if we inject billions of Antibiotic resistance genes into billions of people?
What happens when the injection contains LNPs that can get to your intestines and transform bacteria with these genes?
Why isn't there sequencing QC of every lot given how cheap this is to perform in 2023?
Why QC things when gov is paying & you have a hall pass and a mandate?
Why isn't there sequencing QC of every lot given how cheap this is to perform in 2023?
Why QC things when gov is paying & you have a hall pass and a mandate?
I want thank @JesslovesMJK @TheJikky @SabinehazanMD @pathogenetics @VBruttel @stevenemassey @Fynnderella1 @hyattjn for helpful comments on this project
This molecule wasn't part of the informed consent but each injection likely includes billions of them. 
If you are going to inject the population with antibiotic resistance genes, you would wise to have a strong antibiotic discovery program.
pfizer.com/science/hot-to…
pfizer.com/science/hot-to…
Some question on the Substack.
There are several stones left unturned in the data as I didn’t want to hold up disclosure of this.
1)do the paired end reads imply linear or circular DNA?
There are several stones left unturned in the data as I didn’t want to hold up disclosure of this.
1)do the paired end reads imply linear or circular DNA?
2)what is the strandedness count? Directional libraries should be able to answer this. Might inform if the vector sequence is DNA or RNA or both.
3) the EMA has limits on the dsRNA and they measure this with some ELISA assay which I’m suspect of?
3) the EMA has limits on the dsRNA and they measure this with some ELISA assay which I’m suspect of?
4)error rates in the vector sequence should be lower than in the mRNA as the mRNA goes through a T7 polymerase step with PseudoU and an RT step with PseudoU. There is no PseudoU in the vector.
5)the SV40 in this vector is just a strong promoter. It is not the entire SV40 genome.
5)the SV40 in this vector is just a strong promoter. It is not the entire SV40 genome.
Is the plasmid sequence DNA or RNA?
DNA will provide sequence from both the watson (antisense) and crick (sense) strands.
mRNA will be sense strand (crick).
We used directional libraries so we can count the sense vs antisense strands.
bitesizebio.com/42077/rna-stra…
DNA will provide sequence from both the watson (antisense) and crick (sense) strands.
mRNA will be sense strand (crick).
We used directional libraries so we can count the sense vs antisense strands.
bitesizebio.com/42077/rna-stra…
Download the BAM files and run these 2 samtools scripts on it. This will count the Forward and Reverse strands in the data per base across the contig. 



Now put those in excel and subtract the Forward read count from the reverse read count and chart the results.
What do we see? We see a foward strand bias for half the vector and then a sudden shift to a reverse strand bias at position ~2100
What do we see? We see a foward strand bias for half the vector and then a sudden shift to a reverse strand bias at position ~2100

Thats the exact location of the T7 promoter which initiates synthesis of the mRNA.
This is hard evidence of the vector sequence being DNA and and not an RNA mirage.
The variability in the strandedness in the mRNA region is very interesting? Maybe G4s or other RNA knots?
This is hard evidence of the vector sequence being DNA and and not an RNA mirage.
The variability in the strandedness in the mRNA region is very interesting? Maybe G4s or other RNA knots?

T7 Promoters are notoriously leaky in E.coli. You dont need T7 bacteriophage around to get expression.
ncbi.nlm.nih.gov/pmc/articles/P…
ncbi.nlm.nih.gov/pmc/articles/P…

Rabbit hole-
Where there are plasmids, There is usually Lipopolysaccharides (LPS) from the E.coli the plasmid is replicated in.
These aggregate spike protein.
pubmed.ncbi.nlm.nih.gov/36050806/
pubmed.ncbi.nlm.nih.gov/33295606/
academic.oup.com/jmcb/advance-a…
nature.com/articles/s4138…
Where there are plasmids, There is usually Lipopolysaccharides (LPS) from the E.coli the plasmid is replicated in.
These aggregate spike protein.
pubmed.ncbi.nlm.nih.gov/36050806/
pubmed.ncbi.nlm.nih.gov/33295606/
academic.oup.com/jmcb/advance-a…
nature.com/articles/s4138…
Someone in the recent spaces I was on raised a good question. references welcome.
What is the differential transformation efficiency of LNPs on mammalians vs bacterial cells. They are designed for membranes.
I think the size plays a big role here.
ncbi.nlm.nih.gov/pmc/articles/P…
What is the differential transformation efficiency of LNPs on mammalians vs bacterial cells. They are designed for membranes.
I think the size plays a big role here.
ncbi.nlm.nih.gov/pmc/articles/P…
But you only need 1 in billion to hit and if there is selection there, the plasmid takes off. E.coli doubling time is 30 minutes. We need clinical verification with qPCR for the vector/insert.
Also temperature and environment play a big role in E.coli transformation efficiency. You can find strains that transform at 37C. Add in patient fevers and even naked DNA with no LNPs will transform the bacteria. Important topic raised. It was VSRF space
zymoresearch.com/products/mix-a…
zymoresearch.com/products/mix-a…
That slope in the stranded chart was bugging me. Fixed the bug. Better depiction here. DNA should sense and antisense reads. mRNA only sense strands. SO when you substract Antisense from Sense across the vector you see the T7 promoter at base 2200. The other chatter might be.. 

RNA fragmentation bias or Pseudouridine pause sites.
• • •
Missing some Tweet in this thread? You can try to
force a refresh











