
I need to raise awareness about an important point in #scRNAseq data analysis, which, in my opinion, is not acknowledged enough:
‼️In practice, most cell type assignment methods will fail on totally novel cell types. Biological/expert curation is necessary!
Here's one example👇
‼️In practice, most cell type assignment methods will fail on totally novel cell types. Biological/expert curation is necessary!
Here's one example👇

Last year, together with @LabPolyak @harvardmed, we published a study in which we did something totally awesome: we experimentally showed how a TGFBR1 inhibitor drug 💊 prevents breast tumor initiation in two different rat models!
Here's a detailed thread on this paper:
Here's a detailed thread on this paper:
https://twitter.com/simocristea/status/1600578512578035733
As you can imagine, this is a big thing. Treating tumors is already hard, preventing them is even harder!
Obviously, the most burning question for us then became: what is the drug actually doing to prevent tumor initiation?
Or, what is different in treated vs. control cells?
Obviously, the most burning question for us then became: what is the drug actually doing to prevent tumor initiation?
Or, what is different in treated vs. control cells?

Long story short: we identified a group of cells popping up/expanding after treatment in both strains (ACI & SD)
How do we know these cells are unique to treatment?
Because all other subpopulations were matched, except this one. So these cells are important.
But what are they?
How do we know these cells are unique to treatment?
Because all other subpopulations were matched, except this one. So these cells are important.
But what are they?
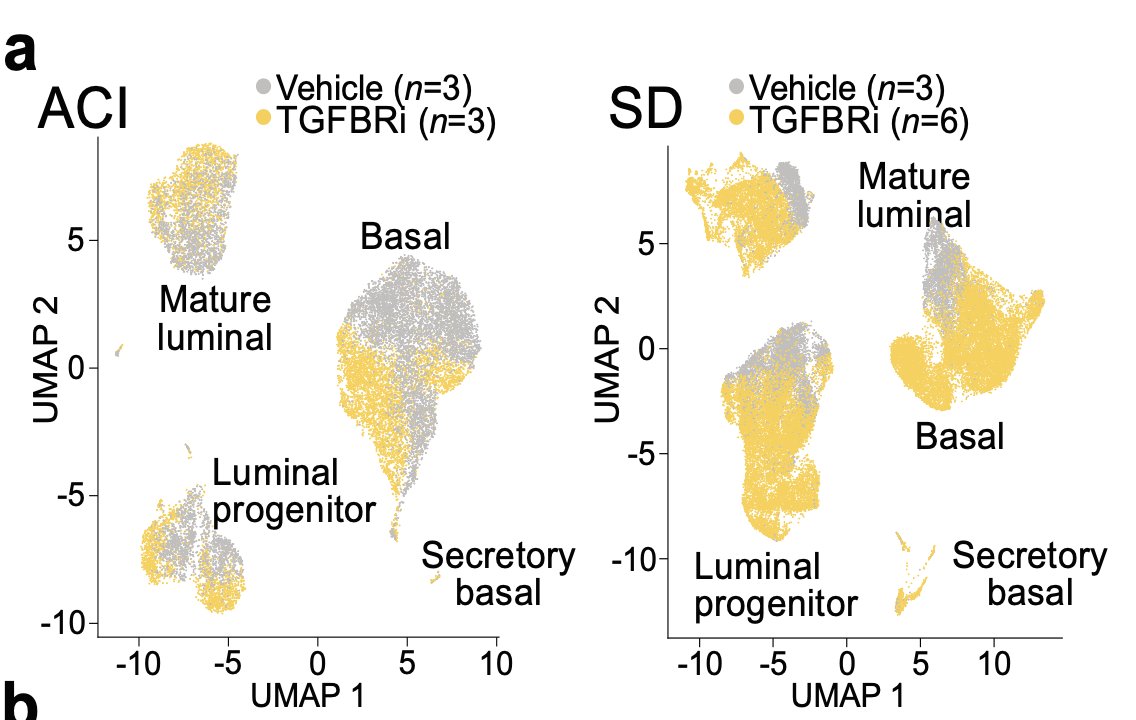
An obvious thing to do is differential expression between these cells and various other relevant groups (s.a. all other cells).
We did that, and got back an interesting list, consisting of many mesenchymal and stem-cell markers, but most of which also characteristic of stroma!
We did that, and got back an interesting list, consisting of many mesenchymal and stem-cell markers, but most of which also characteristic of stroma!

‼️All cell type assignment methods we tried failed to characterize this population accurately. Most of them labeled it "stroma".
But, we weren't easily fooled.
@nellage has *tens of years* of experience with this experimental model & I spent *years* researching relevant papers.
But, we weren't easily fooled.
@nellage has *tens of years* of experience with this experimental model & I spent *years* researching relevant papers.
Our teams spent >2 years investigating these cells.
We discussed hundreds of hours about them.
We embarked on costly & time-consuming experiments to dig deep.
(Science obsession at its best🤫)
All because we wanted to know for ourselves: is this really a new epithelial type??
We discussed hundreds of hours about them.
We embarked on costly & time-consuming experiments to dig deep.
(Science obsession at its best🤫)
All because we wanted to know for ourselves: is this really a new epithelial type??
So, how did we decide if this population is novel epithelium or stroma?
Two main strategies:
1. We knew the literature had evidence for extremely rare (<0.1%) progenitor mammary populations related to tumor initiation. We found lots of similarities with those populations.
Two main strategies:
1. We knew the literature had evidence for extremely rare (<0.1%) progenitor mammary populations related to tumor initiation. We found lots of similarities with those populations.

‼️ 2. We actually did experiments to validate the epithelial (and not stroma!) nature of these cells. We found rare cells in the breast with both epithelial markers, as well as part of this subpopulation.
These experiments were tough because of the rarity of these cells.
These experiments were tough because of the rarity of these cells.

All in all, we gathered substantial evidence (both computational & ultimately experimental) that this population is indeed a novel epithelial type, and not just stroma.
Why did then *none* of the multiple tools we applied signal the novel nature of this subpopulation?
Why did then *none* of the multiple tools we applied signal the novel nature of this subpopulation?
When thinking about it, it's actually pretty straight-forward why.
There's absolutely no magic: cell assignment tools need references to match cells to, and assign based on max similarity with references.
Obviously, with a novel cell type, there's no reference to match it to.
There's absolutely no magic: cell assignment tools need references to match cells to, and assign based on max similarity with references.
Obviously, with a novel cell type, there's no reference to match it to.
Still, in such situations (unmatched to references), most methods claim to at least flag novel subpopulations.
But how about intermediate populations transitioning between cellular types, with context-dependent roles?
How about subpopulations very similar to other cell types?
But how about intermediate populations transitioning between cellular types, with context-dependent roles?
How about subpopulations very similar to other cell types?
The truth is that, in such cases, cell type assignment tools will fail, almost by definition. This expected behavior shouldn't surprise us.
This is why expert/biological knowledge is necessary & has authority over any algorithm
Also, biological validation is the ultimate proof!
This is why expert/biological knowledge is necessary & has authority over any algorithm
Also, biological validation is the ultimate proof!
‼️ I want to make it explicit that I am not claiming cell type assignment algorithms are not performing well.
I think such algorithms are nothing short of extraordinary.
It's just that they can't do all the work for us.
We also need to understand the biology behind our data.
I think such algorithms are nothing short of extraordinary.
It's just that they can't do all the work for us.
We also need to understand the biology behind our data.
The reason I am bringing this up is that, in my experience, it comes up repeatedly during discussions over concrete #scRNAseq datasets.
I've seen many #Bioinformatics analysts somehow reluctant to "override"/question the assignments of an algorithm.
That is missing the point.
I've seen many #Bioinformatics analysts somehow reluctant to "override"/question the assignments of an algorithm.
That is missing the point.
As scientists, every decision we take needs to be justified.
Once justified and backed-up by evidence, it is valid.
Once the point is valid, it doesn't matter if algorithm X or method Y say differently.
We only want one thing: our claim to be TRUE, to the best of our knowledge
Once justified and backed-up by evidence, it is valid.
Once the point is valid, it doesn't matter if algorithm X or method Y say differently.
We only want one thing: our claim to be TRUE, to the best of our knowledge
‼️Finally, actionable:
Understanding how cell type assignment algorithms actually work helps us also understand their limitations.
That's why my advice to #Bioinformatics folks is to read the papers behind *all* the algorithms they are applying.
(Sorry, no exceptions allowed!)
Understanding how cell type assignment algorithms actually work helps us also understand their limitations.
That's why my advice to #Bioinformatics folks is to read the papers behind *all* the algorithms they are applying.
(Sorry, no exceptions allowed!)
• • •
Missing some Tweet in this thread? You can try to
force a refresh










