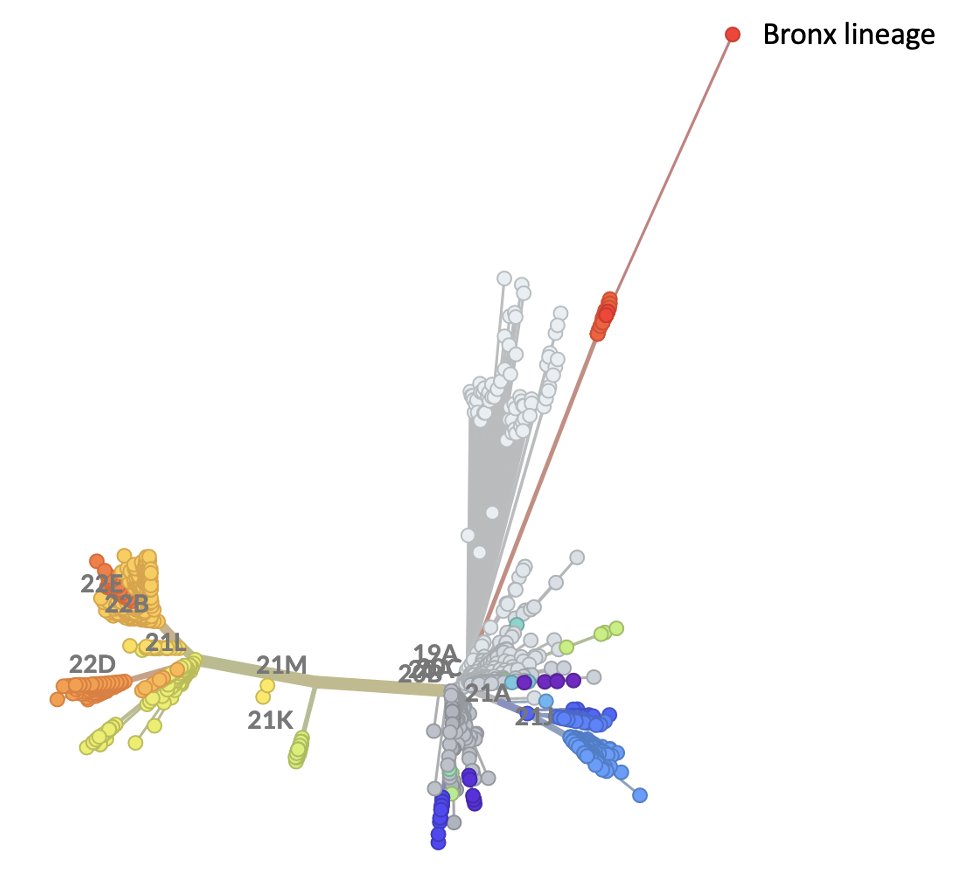
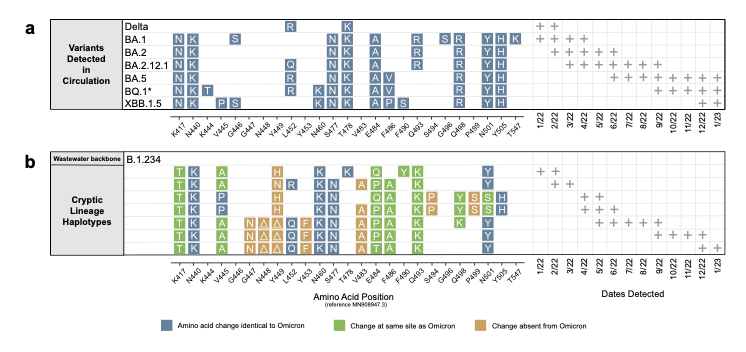
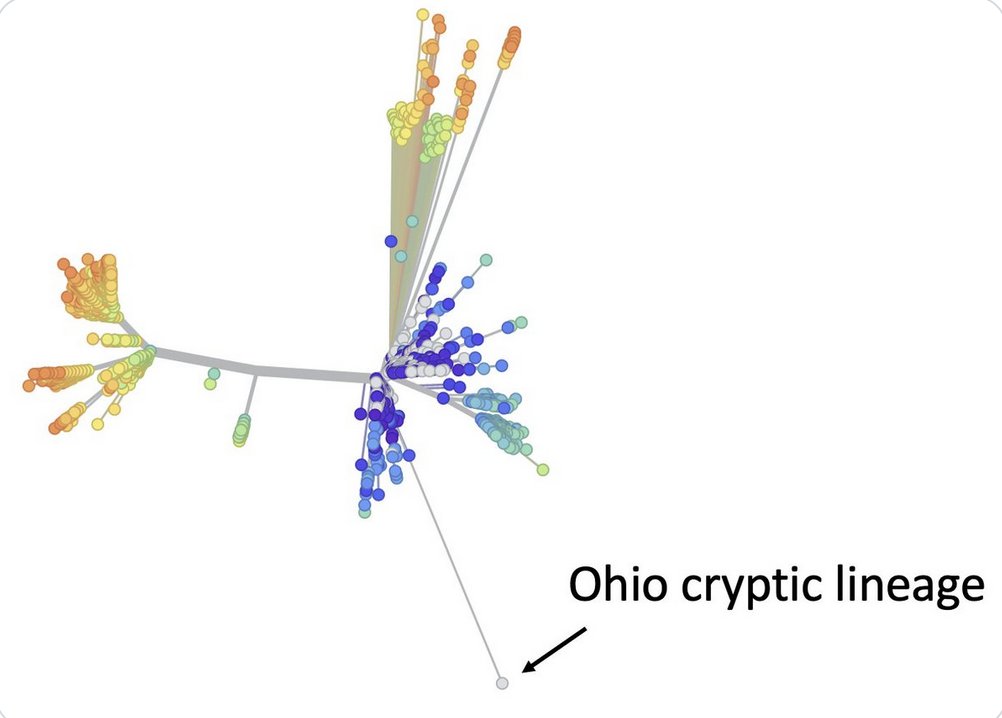
Ohio cryptic lineage update.
We’ve made no progress identifying the individual, but we have learned a few things.
1/
We’ve made no progress identifying the individual, but we have learned a few things.
1/

First, the signal is almost always present in the Columbus Southerly sewershed, but not always at Washington Court House. I assume this means the person lives in Columbus and travels to WCH, presumably for work.
2/
2/

Second, the signal is increasing with time. Washington Court House had its highest SARS-CoV-2 wastewater levels ever in May, and the most recent sequencing indicates that this is entirely the cryptic lineage.
3/
3/

Third, I’ve tried to calculate how much viral material this person is shedding. (Multiply the cryptic concentration by the total volume). I’ve done this several times and gotten pretty consistent results.
They are shedding a few trillion (10^12) genomes/day.
4/
They are shedding a few trillion (10^12) genomes/day.
4/
What does this tell us? How much tissue is infected? It’s impossible to know for sure. Chronically infected cells probably don't release much, but acutely infected cells produce a lot more. I gather a typical output in the lab is around 1,000 virus per infected cell.
5/
5/
If we assume we are getting 1,000 viral particles per infected cell, that would mean there are at least a billion infected cells. The density of monolayer epithelial cells is around 300k cells/sq cm. A billion cells would represent around 3.5 square feet of epithelial tissue!
6/
6/
Don’t get me wrong. The intestines have a huge surface are and 3 square feet is a tiny fraction of the total.
But it’s still a massive infection, no matter how you slice it.
7/
But it’s still a massive infection, no matter how you slice it.
7/
I do not know of any persistent infections that shed this much virus without killing the patient. (Correct me if I’m wrong). The closest would probably be HCV, an infection that often ends in liver cancer.
8/
8/
My point is that this patient is not well, even if they don’t know it, but they could probably be helped if they were identified.
9/
9/
How do you figure out if you have a COVID GI infection. Most rapid antigen test are not explicitly for stool. However, I know of at least one that is. The instructions from that test say to mix a ‘matchhead’ worth of feces with the buffer.
10/
dyonmedical.com/en/corona-ag-a…
10/
dyonmedical.com/en/corona-ag-a…

I can tell you that with very positive wastewater samples we got a positive by just sticking the swab into the wastewater before adding it to the buffer. Either way, it is essential to use the buffer provided or the test doesn’t work.
11/
11/
If you do get a positive reading, I would suggest having someone else try with the same kind of test with their feces to be sure it isn’t a false positive. Weird shit happens.
12/
12/
If you are the individual, let me know. There is a lab in the US that can do 'official' tests for COVID in stool, and there are doctors that I can put you in contact with that would like to try to help you.
13/
13/
What's more, if we can figure out how these infections occur, and how to treat them, it could potentially help out a whole lot of people.
14/
14/

Two comments I should have made in my thread.
Washington Court House is a town of 15k (not an actual courthouse).
I haven't invaded anyone's privacy. Approximately 1600 people commute between Columbus and WCH. That isn't enough information to pinpoint the source.
Washington Court House is a town of 15k (not an actual courthouse).
I haven't invaded anyone's privacy. Approximately 1600 people commute between Columbus and WCH. That isn't enough information to pinpoint the source.
• • •
Missing some Tweet in this thread? You can try to
force a refresh

 Read on Twitter
Read on Twitter