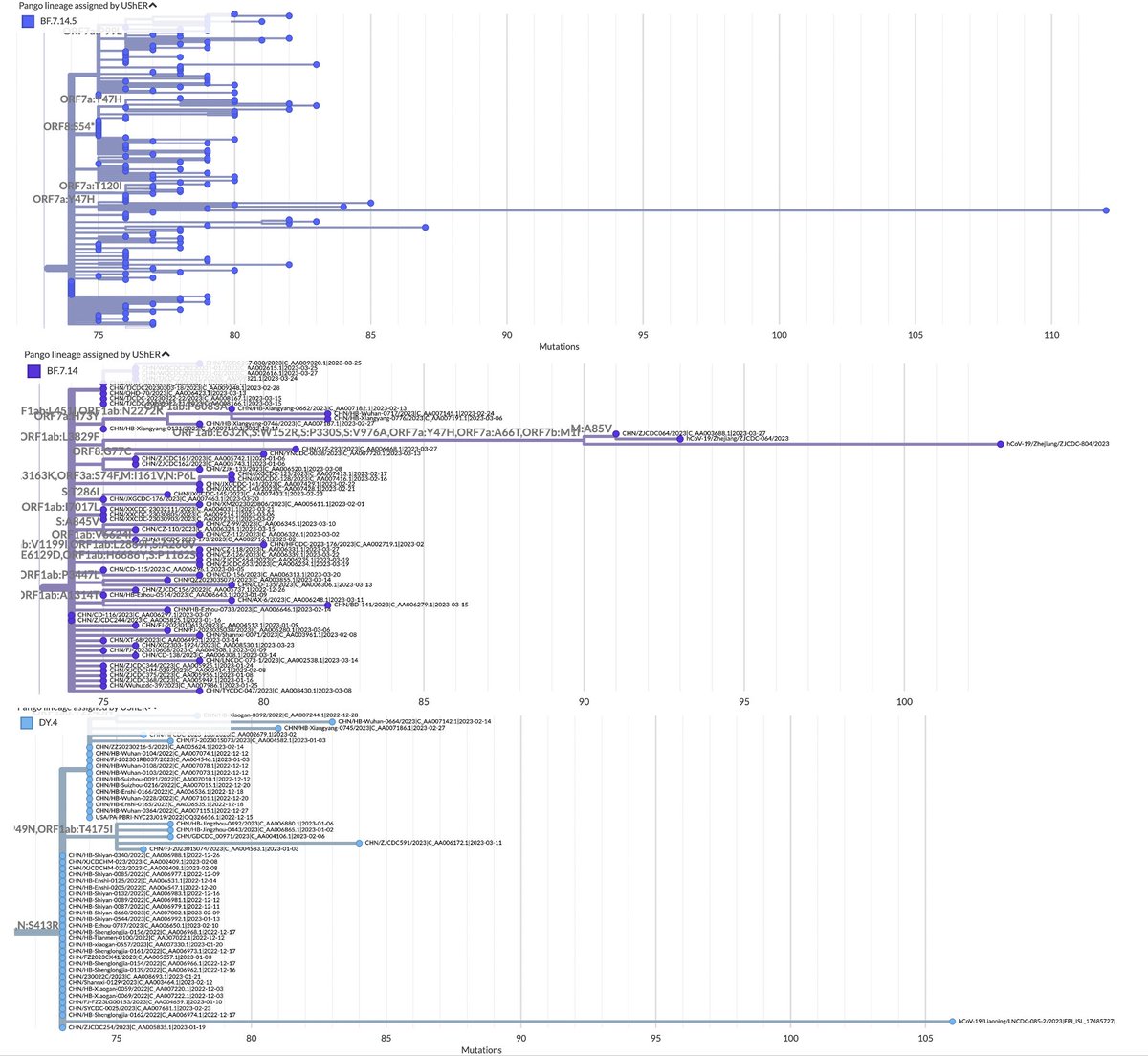
What's going on with ORF7b? Something unusual has occurred in ORF7b in two unrelated variants—two of the only non-XBB variants that remain competitive. They took entirely different routes to reach the same end point: a new ORF7b protein.
But first a short primer on ORF7b. 1/
But first a short primer on ORF7b. 1/
SARS-CoV-2 genes are often divided into three categories:
1. Non-structural proteins (NSPs)
2. Structural proteins (spike, envelope, membrane, nucleocapsid or S, E, M, N)
3. Accessory proteins 2/
1. Non-structural proteins (NSPs)
2. Structural proteins (spike, envelope, membrane, nucleocapsid or S, E, M, N)
3. Accessory proteins 2/
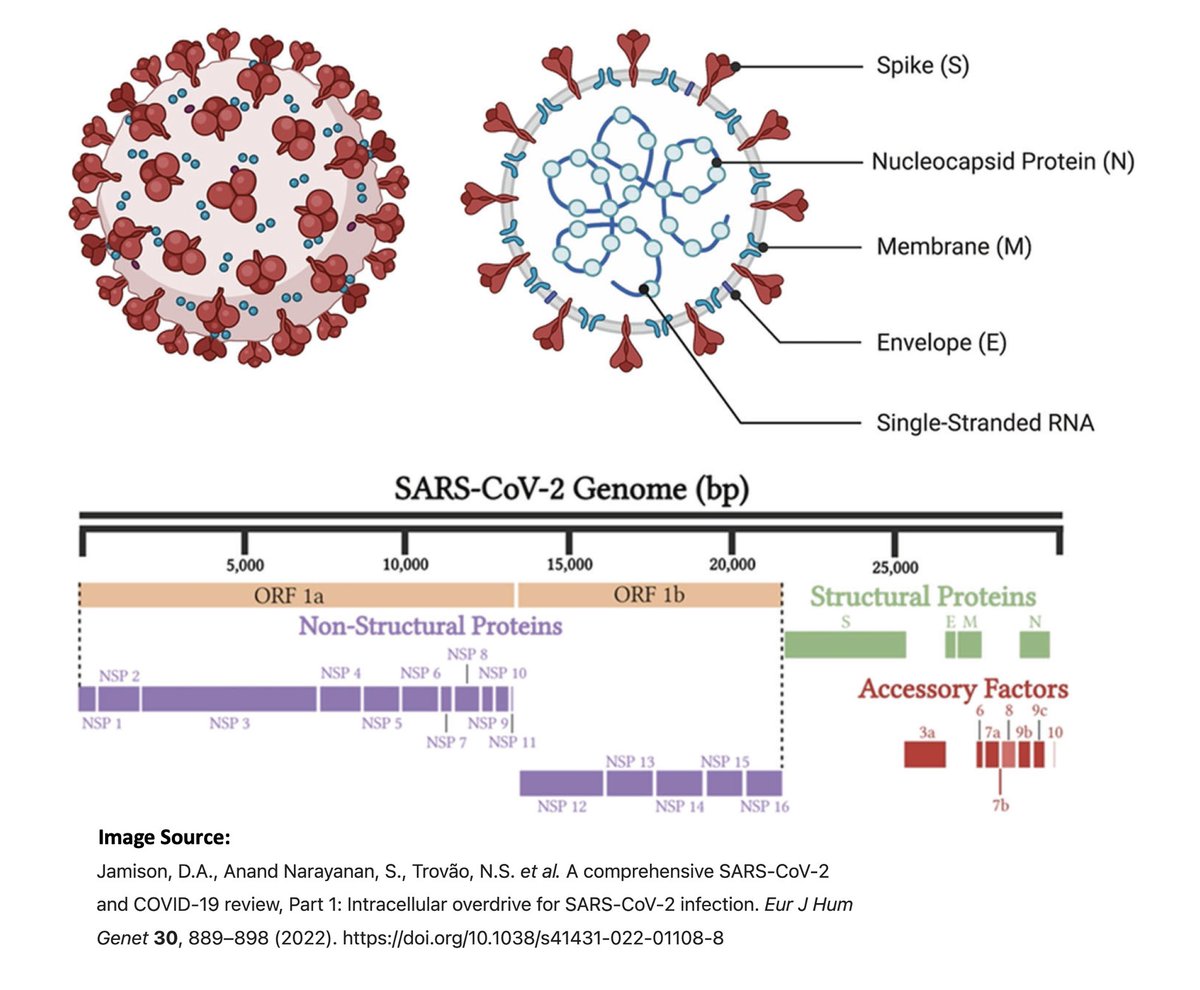
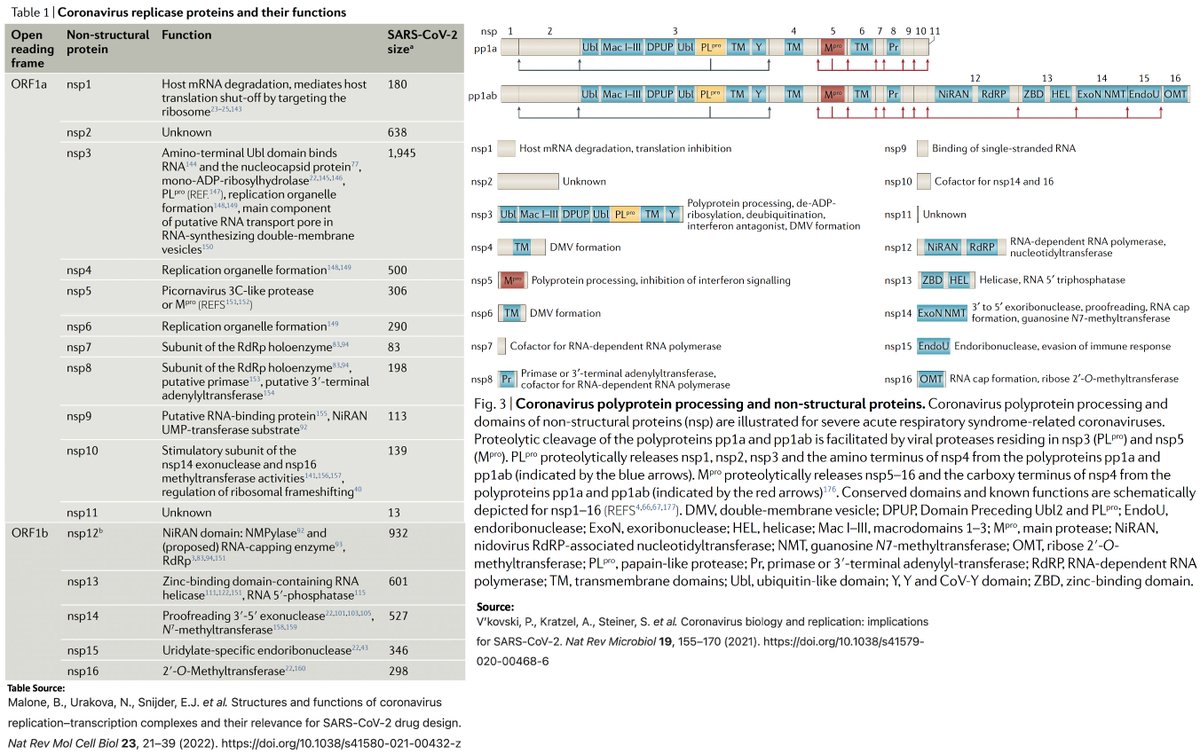
The 16 NSPs are in ORF1ab & make up the first ~2/3 of the genome. They have diverse functions, but most are primarily involved in viral replication in one way or another. Here are descriptions of the functions of the 16 NSPs from a few different sources. 3/ 


Structural proteins (envelope, membrane, spike, nucleocapsid—E, M, S, N) compose the physical structure of the virus. N has also been implicated in innate immune evasion.
4/
4/
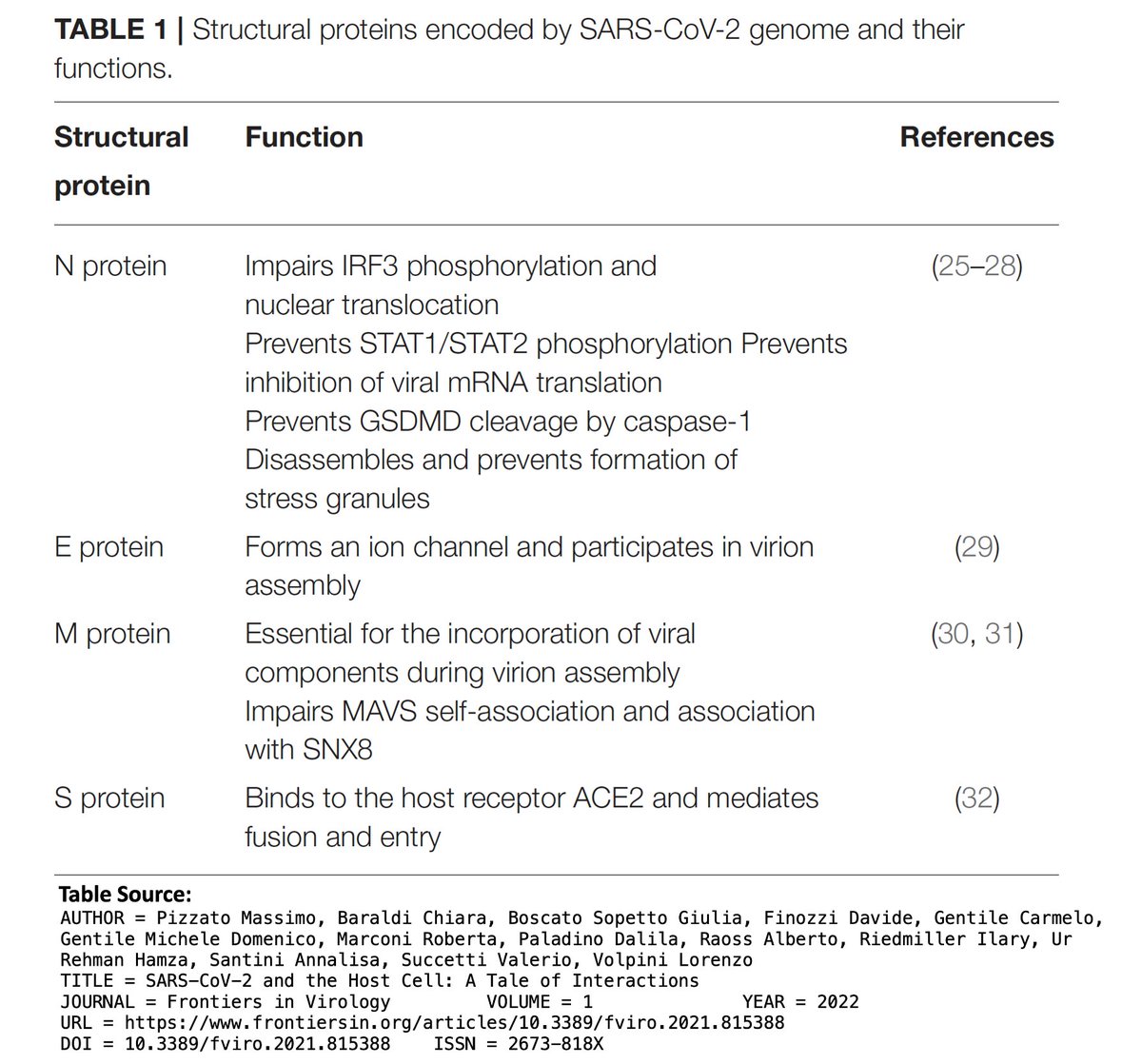
Accessory proteins (ORF3a, ORF6, ORF7a, ORF7b, ORF8, ORF9b, & possibly others) are less essential than the first two categories, & seem to be mainly involved in immune evasion. 5/ 

But it's not always clear what accessory proteins do. ORF8, after having had dozens of powerful functions attributed to it, has been nearly erased, resulting in an apparent increase in fitness. ORF7a has also been destroyed repeatedly. 6/
https://twitter.com/LongDesertTrain/status/1645223059698139142
ORF7b is the smallest protein, at 43 amino acids (AA), & early ORF7b stop codons, which prematurely halt protein construction, have made regular appearances (though not as commonly as in ORF8). At first, the new ORF7b mutations seem similarly destructive of ORF7b. 7/ 
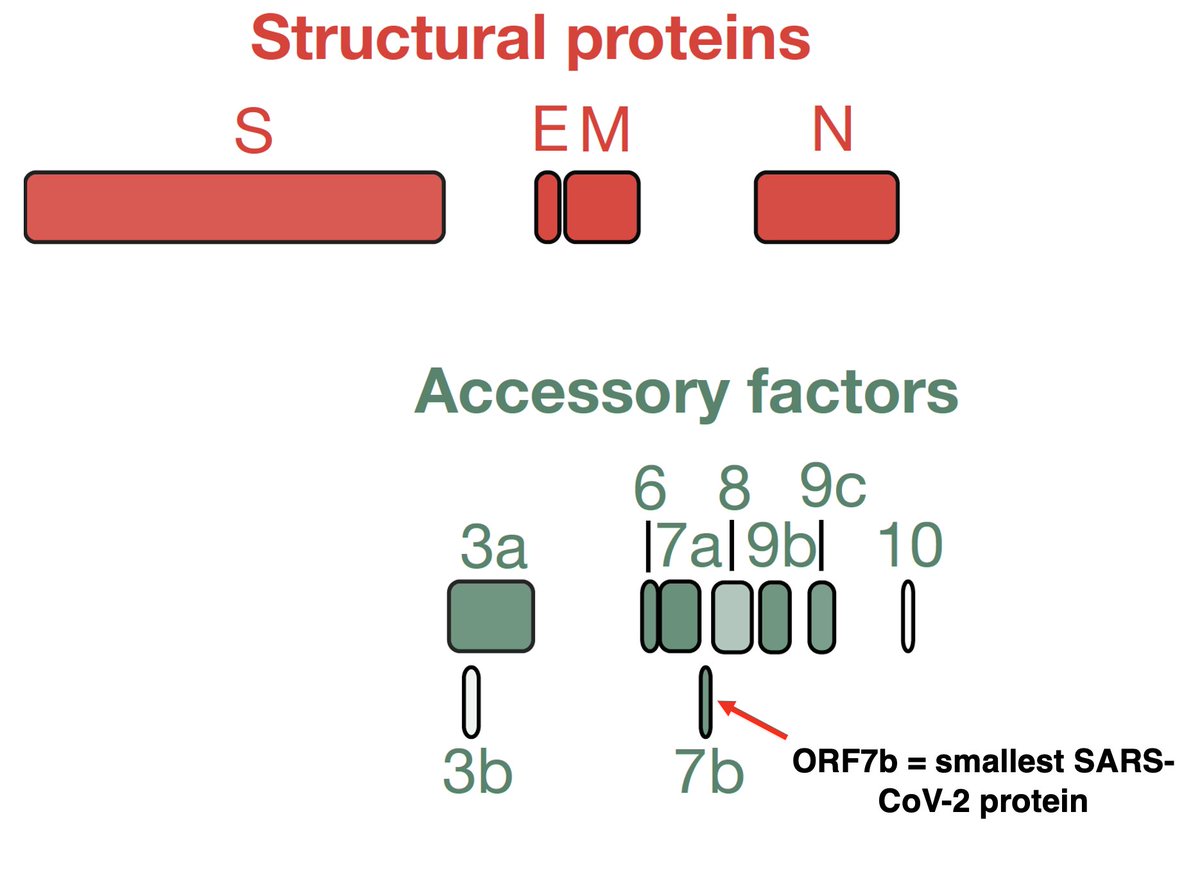
Each AA is coded by a "codon" made up of 3 nucleotides (nt), so any deletion not divisible by 3 completely changes the composition of the AA appearing after it. This is called a frameshift. Example: a 1-nt deletion in ORF6 in XBB.1.28.1 (spotted by @shay_fleishon). 8/ 
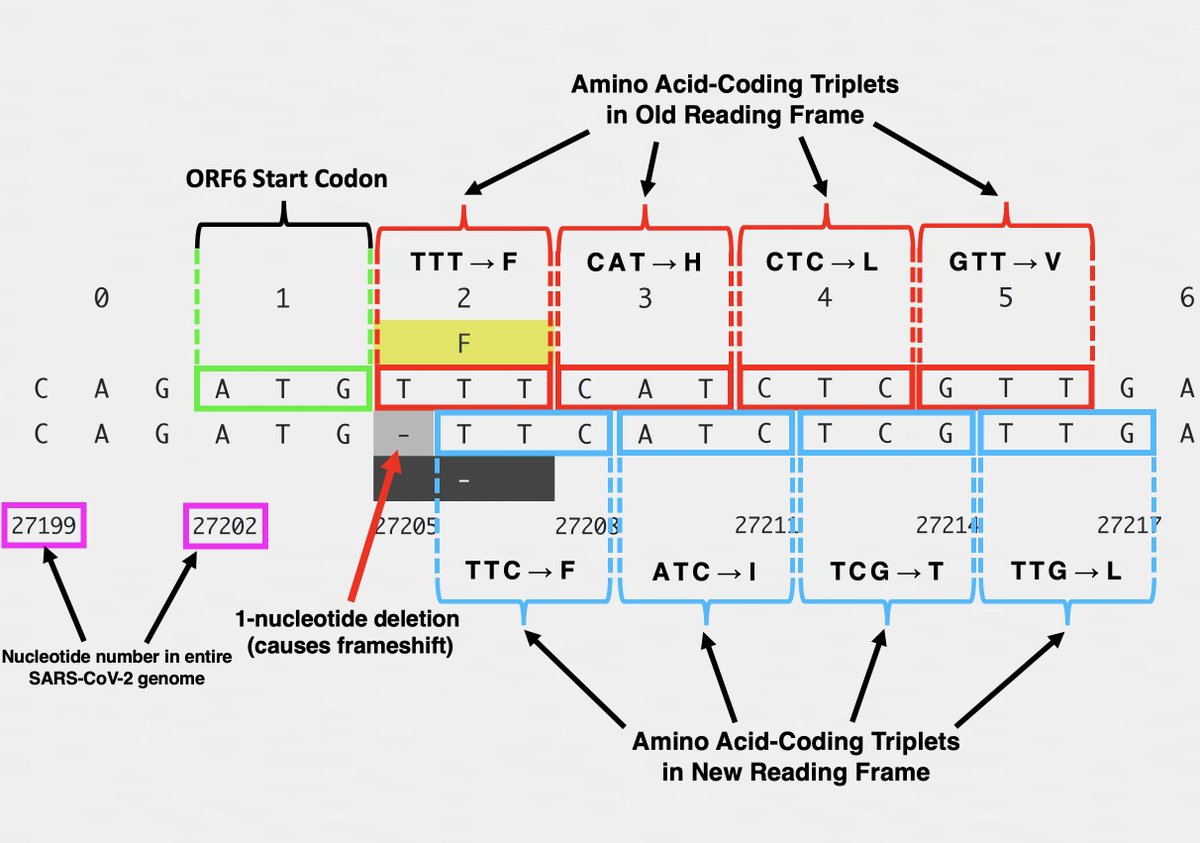
Frameshifts usually quickly lead to a stop codon, truncating the protein & making it incapable of carrying out its previous function. The 1-nt deletion in ORF6 in XBB.1.28.1, for example, finds a stop codon at AA site #11, hacking off the last 84% of ORF6. 9/ 
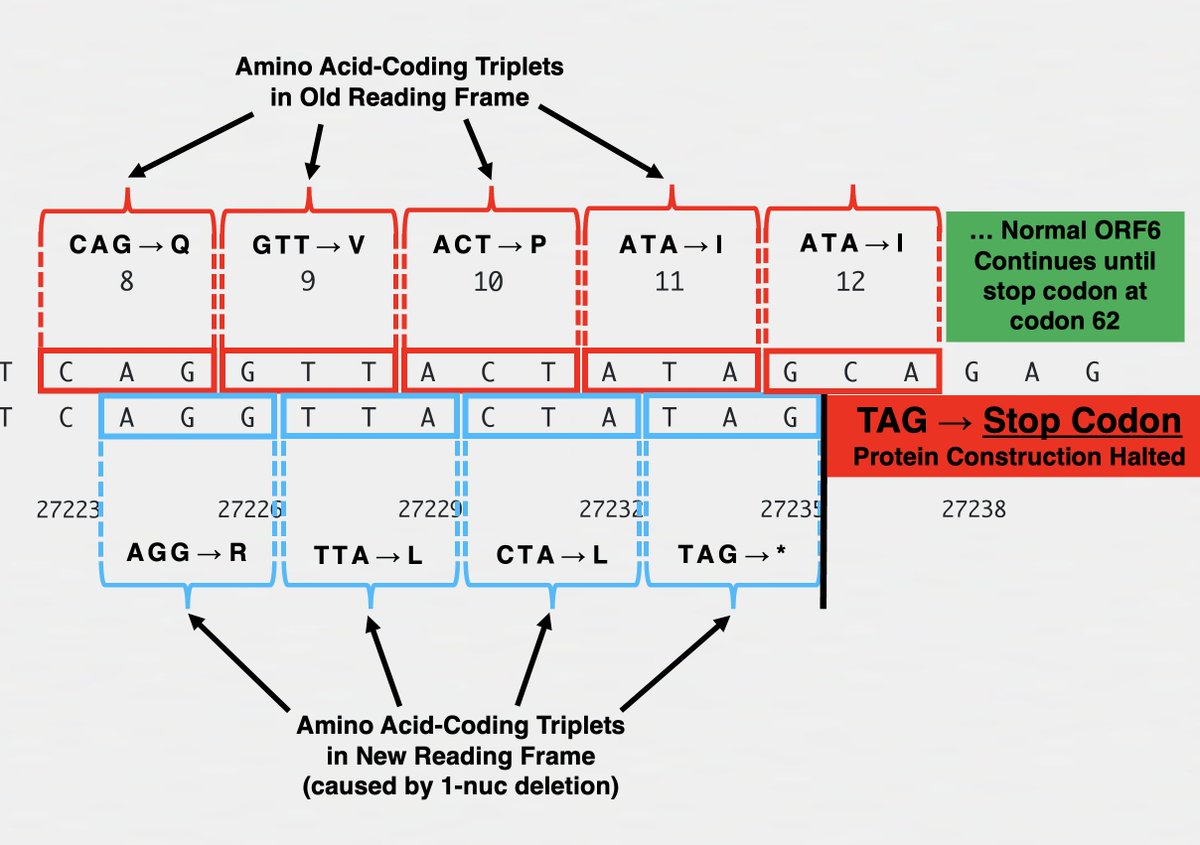
One variant—XBC.1.6, a Deltacron—has a 2-nucleotide deletion in ORF7b codon 13, causing a frameshift. But this frameshift doesn't lead to a stop codon until codon 32-33. 10/ 
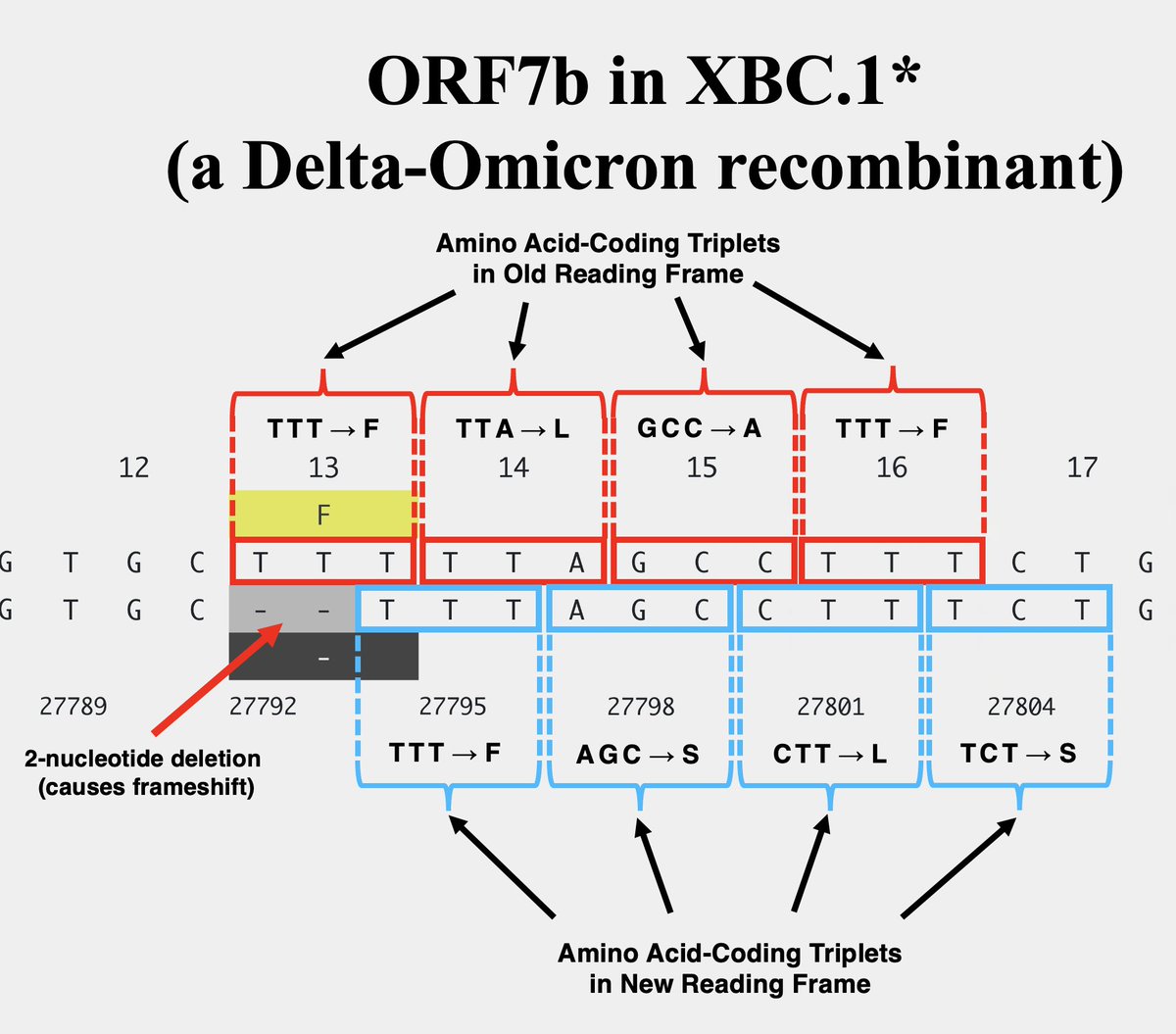
The other variant—FR.1, a BN.1 descendant (which is a BA.2.75 descendant)—has a 1-nucleotide insertion at the same location. This causes a frameshift *identical* to the 2-nuc deletion in XBC.1.6, the only difference being an extra F residue in FR.1. 11/ 
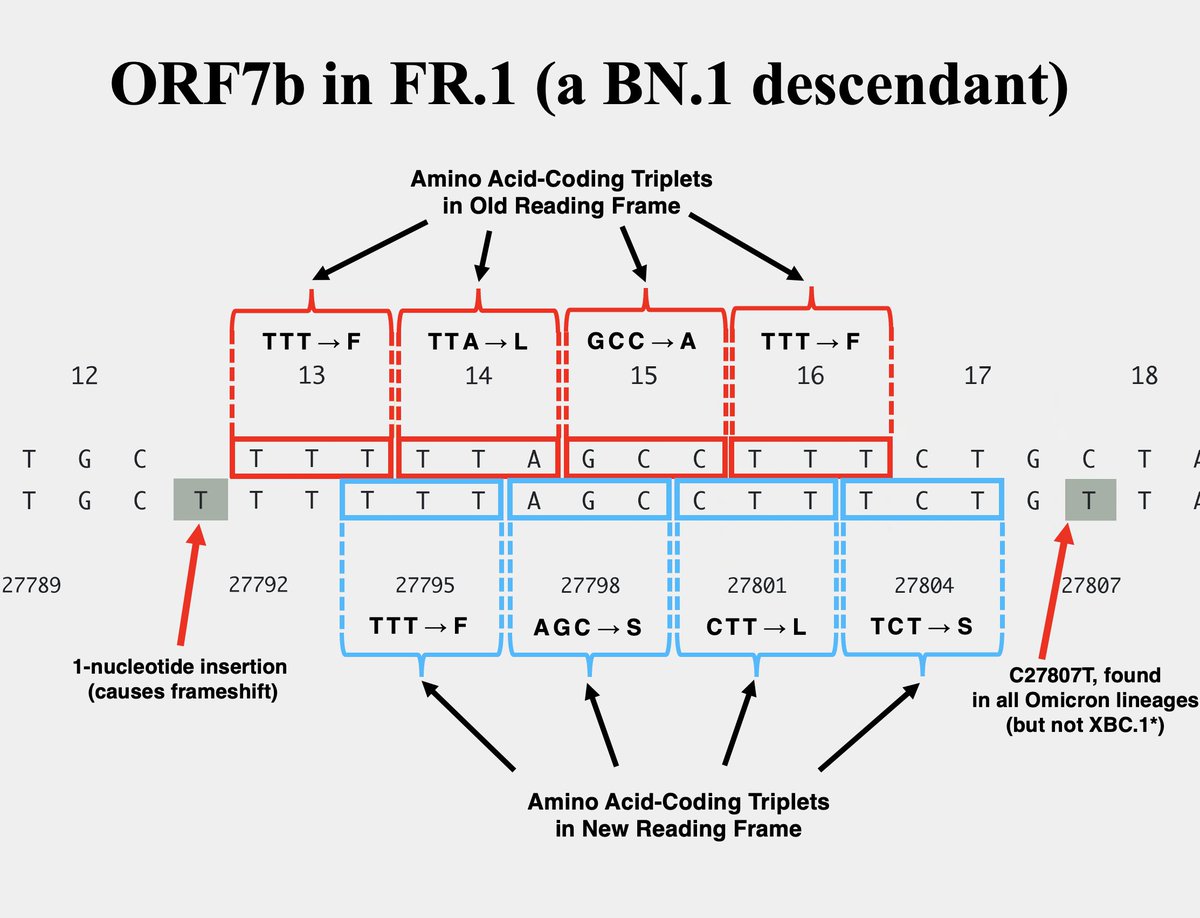
So the new, frameshifted ORF7b is about ~75% as long as the old version. But the amino acid composition drastically differs, particularly in the number of polar vs nonpolar AA residues. Post-frameshift, 7 of the first 14 AA are polar vs 0/14 in the old ORF7b. 12/ 
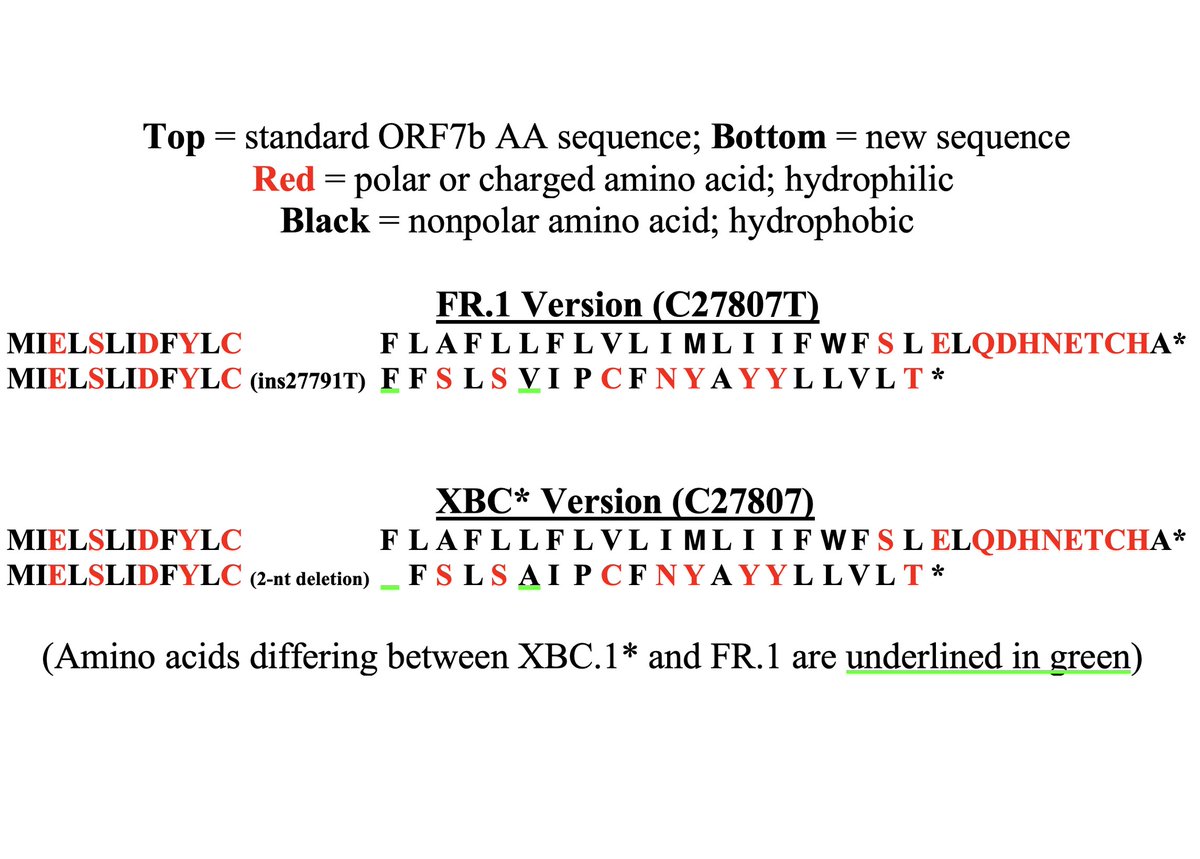
But 10 of the last 11 AA residues of the original ORF7b are polar/hydrophilic, & the new, truncated ORF7b lacks these altogether. I think it's safe to say these are two very different proteins. 13/ 

But what does it all mean? The fact that two of the most competitive non-XBB variants have both stumbled on the same new protein through different routes suggests it may be beneficial in some way, perhaps throwing a spanner in the works of our immune response. 14/
Or the new ORF7b could be a junk protein w/no function. ORF7b likely isn't important enough to have a major impact. But I think this illustrates the sort of evolutionary surprises that are possible. Even as some are erased (ORF8), entirely new proteins can be created. 15/
Indeed at least one already has! It resulted from a 3-nucleotide mutation in N, creating a new TRS-B & leading to the production of the N* protein, which has been shown to interfere with the immune response and increase viral loads. 16/ nature.com/articles/s4146…
N* is now universal except for in Deltacrons like the XBC lineage. It's was a major development that hasn't seemed to get the attention it merits. But that would be an entirely different thread. Time to end this one. 17/
https://twitter.com/KroganLab/status/1583854636770816000
As always to the labs, lab workers, and researchers doing the groundwork, without which none of this would be possible. As funding is reduced and sequencing decreases, their work is more important than ever. 18/end
• • •
Missing some Tweet in this thread? You can try to
force a refresh

 Read on Twitter
Read on Twitter