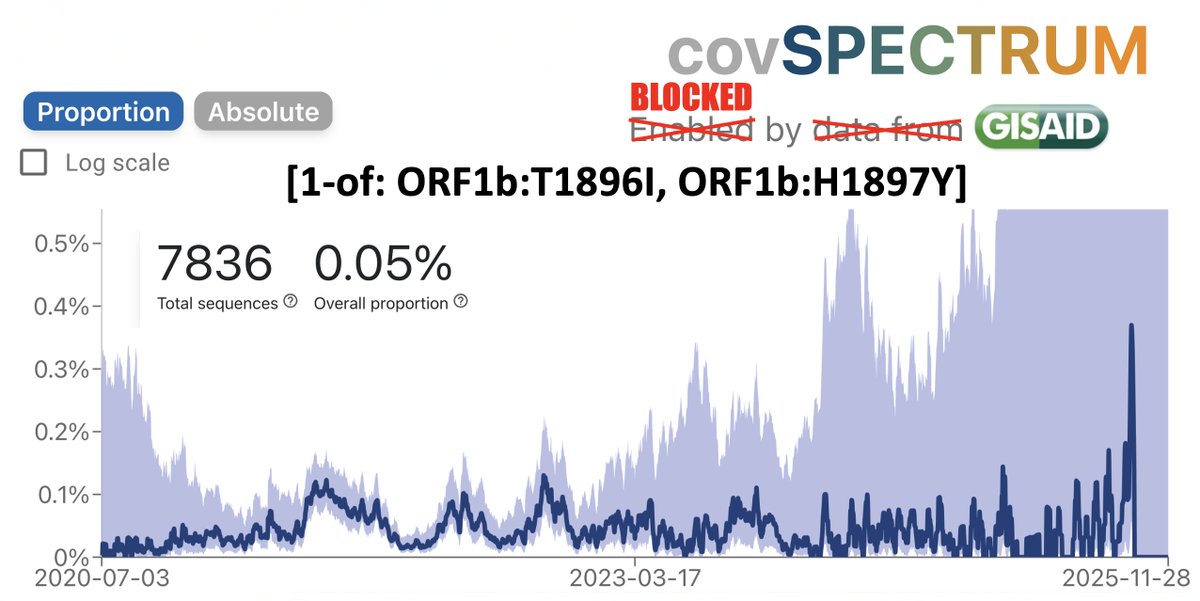
How important is ORF9b? More important, I think, than almost anyone has realized. ORF9b is often neglected—it is ignored on phylogenetic trees & on GISAID sequences. I see 9 lines of evidence indicating it's a major player. But they require some background. /1 
ORF9b is encoded by a stretch of genome that entirely overlaps w/the nucleocapsid (N) gene. But ORF9b is "out of frame" with respect to N, so the amino acids they code for are totally different, as I've tried to depict below. 2/ 

ORF9b is an accessory protein (as are ORF3a, ORF6, ORF7a, ORF7b, & ORF8). As explained in a previous thread, accessory proteins appear to be involved in immune evasion but are less essential to viral replication other proteins. 3/
https://twitter.com/LongDesertTrain/status/1666220308599037955
All SARS-CoV-2 accessory proteins (along with the structural proteins S, E, M, & N) are encoded by subgenomic mRNAs. The way these sgmRNAs are transcribed (copied) is very strange & not particularly efficient. I'll try to explain how it works. 4/
The SARS-CoV-2 genome is a single strand of (+)-sense RNA, ~30,000 nucleotides long. (+)-sense means ribosomes (a cell's protein-making machines) can directly translate it into proteins. (-)-sense RNA viruses like influenza must copy their (-)RNA into +RNA first. 5/ 

A protein called the RdRp (or NSP12), in concert with several other proteins, carries out all transcription (copying) of SARS-CoV-2 RNA. It copies the genome "backwards," from the end to the beginning (right to left in diagrams in this thread). 6/ 

The RdRp is a magical little machine that zips along the genome, & w/astonishing accuracy, pairs every single nucleotide with its correct partner, making a (near-)perfect copy. It doesn't seem like this magic act should be possible, but there's a lot in biology like that. 7/ 

In normal genome replication, the RdRp runs from one end of the genome to the other, copying the whole thing into a (-)-sense genome, which is later copied back into a (+)-sense genome, which is either packaged in new viruses or fed into a ribosome to make proteins. 8/ 

But the full genome can only make non-structural proteins (NSPs) encoded by ORF1ab. It cannot produce any of the structural proteins (S, E, M, N) that make up the physical virus or any accessory proteins (3a, 6, 7a, 7b, 8, 9b). 9/ 

So how are the other proteins made? It's extremely weird. No one would ever design a system this way. But evolution is not an engineer. It's a sleepwalker. 10/
Science itself is something of a sleepwalker. Novelist Arthur Koestler wrote a great book on the history of physics called The Sleepwalkers. Much of it focuses on Johannes Kepler—sleepwalker scientist extraordinaire, featured in this previous thread. 11/
https://twitter.com/LongDesertTrain/status/1515090370086031367
Here's how it works: As the RdRp proceeds backwards along the (+)-sense genome, copying as it goes, it runs into stretches of genome called TRS's—transcription regulation sequences. Each TRS occurs before (left of) one protein-encoding sgRNA. Image via @jbloom_lab paper 12/ 

Most of the time, the RdRp skips over a TRS, so nothing happens. But a small percentage of the time, when the RdRp crosses a TRS, it pulls off another miraculous trick: it JUMPS off and SKIPS to the start of the genome. 13/ 

After this astounding jump, RdRp finishes by copying the first ~65 nucleotides in the genome. The result is what's called "subgenomic mRNA" (sgmRNA)—it has both ends of the full genome, but is missing 70-95% in the middle, including all of ORF1ab. 14/ 

There are 9 basic forms of sgmRNA. They're "nested" because each is contained in the next larger sgmRNA, which is contained in the next larger, etc., like Matryoshka dolls. They float in the cell until they meet a ribosome, which makes 1—and only 1—protein from each. 15/ 

This is pretty inefficient. In order to make the spike protein, for example, the RdRp copies a stretch of genome that includes not only spike, but every sgmRNA after spike as well, none of which are translated into proteins. That's a lot of wasted resources. 16/ 

Why does the RdRp make that crazy jump to the start of the genome just to copy a tiny stretch (the first 70 nucs)? Why not just jump off at a TRS and cut the resulting sgmRNA loose so it can find a ribosome & be translated into a protein? /17 

Human ribosomes won't translate just any RNA. It requires certain features on both ends. The start & end of the SARS-CoV-2 genome imitate features of human mRNA, including a "cap," RNA structures, & poly-A tail. Without these end-features, no sgRNA produces a protein. 18/ 

What exactly *is* a TRS, which causes the RdRp to make these jumps? It's a simple sequence of nucleotides. The core sequence is AAACGAAC. This is the exact sequence found in the location at the start of the genome—called the TRS-L—where the RdRp jumps to. 19/ 

The nucleotides being strung together by the RdRp at a TRS therefore match up with the TRS-L nucleotides near the start of the genome and will tend to stick to them if brought close, after which the RdRp copies the remaining ~65 nucs. 20/ 

Though the core sequence is AAACGAAC, if a TRS also has nucleotides that match those to the left or right the TRS-L, it works even better, & more of its sgmRNA will be produced. This is called "extended homology."
*END Tutorial Section of thread* 21/
*END Tutorial Section of thread* 21/

Now that that's out of the way, we can discuss the nine signs that I think indicate ORF9b is a very important protein. The first 3 could be interpreted to show the importance of N or ORF9b, but the last six are specific to ORF9b. First in list form. 22/
1) End-of-ORF8 extended homology
2) Two overlapping TRS's
3) Gamma insertion = 3x TRS
4) Additional TRS's + early in-frame start codons + ORF9b:P10F
5) Alpha's 3-nuc N:D3L
6) Delta & Alpha's ∆A28271
7) Omicron's A28271T
8) The I5T advantage
9) ORF9b convergence in chronics 23/
2) Two overlapping TRS's
3) Gamma insertion = 3x TRS
4) Additional TRS's + early in-frame start codons + ORF9b:P10F
5) Alpha's 3-nuc N:D3L
6) Delta & Alpha's ∆A28271
7) Omicron's A28271T
8) The I5T advantage
9) ORF9b convergence in chronics 23/
1) End-of-ORF8 Extended Homology
ORF9b & N share a TRS that starts at the tail end of ORF8. The "AA" in the core AAACGAAC motif overlaps w/the ORF8 stop codon. Even in wild-type SARS-CoV-2, there are already three nucleotides of extended homology… 24/
ORF9b & N share a TRS that starts at the tail end of ORF8. The "AA" in the core AAACGAAC motif overlaps w/the ORF8 stop codon. Even in wild-type SARS-CoV-2, there are already three nucleotides of extended homology… 24/

But the virus clearly wants more. Two of the most common mutations—evolving independently, again & again, in a multitude of lineages—create additional extended homology for the ORF9b/N TRS, matching the TRS-L even better & increasing N/ORF9b production. 25/ 

This extended homology for the ORF9b/N TRS appears to come in numerous forms (see 5 below), but I suspect these are mostly different interpretations of one or two underlying patterns that in reality form through recombination with the TRS-L. /26 

2) The Two Overlapping ORF9b/N TRS's
I'm not sure if this has been noticed or written about elsewhere, but there are pretty clearly *two*, overlapping TRS's for ORF9b/N. The second one is slightly suboptimal but only slightly. /27
I'm not sure if this has been noticed or written about elsewhere, but there are pretty clearly *two*, overlapping TRS's for ORF9b/N. The second one is slightly suboptimal but only slightly. /27

The AACAAAC in the second ORF9b/N TRS is nearly ideal. The middle A (just after C), is G in the ideal TRS form, but A is a pretty good replacement for G (for reasons that would require a separate thread to explain). /28 

This double, overlapping TRS has gotten little or no attention, but it's unique to the ORF9b/N TRS & seems another sign of the vital importance of ORF9b/N. And another, remarkable phenomenon that, IMO, seals the case for the functionality of the double TRS… /29 

3) Gamma's AACA Insertion Creates a *Triple* TRS for ORF9b/N
I didn't start analyzing genomes until a year after Gamma disappeared from circulation, so I only noticed this recently. Gamma has a 4-nuc insertion within the double TRS that creates a *triple,* overlapping TRS. 30/
I didn't start analyzing genomes until a year after Gamma disappeared from circulation, so I only noticed this recently. Gamma has a 4-nuc insertion within the double TRS that creates a *triple,* overlapping TRS. 30/

Furthermore, this third TRS is even better than the 2nd overlapping, nearly ideal TRS. It's AAACAAAC—almost identical to the ideal AAACGAAC—and since A is a good replacement for G, this one is very nearly perfect. /31 

Gamma always seemed to compete better & cause more reinfections than you'd expect based on its modest antibody-evasion capability. Perhaps this triple ORF9b/N TRS is part of the reason. ORF1a:K1795Q is another candidate. 32/
https://twitter.com/SolidEvidence/status/1668995927258849280
Before going to #4-7, a brief explanation is required. N & ORF9b share a TRS, meaning the sgmRNA is identical for both. But N's start codon comes before ORF9b's, so any ribosomes encounter it first & should translate N, not ORF9b. Then how does ORF9b exist at all? /33 

Some start codons are occasionally bypassed by ribosomes. This is called "leaky scanning." When the N/ORF9b sgmRNA enters a ribosome, the N start codon is bypassed a small percentage of the time, resulting in ORF9b protein production. /34
4) Additional Suboptimal TRS's + Early In-Frame Start Codons Early in ORF9b + ORF9b:P10F
Because N's start codon precedes ORF9b's, ribosomes scanning their shared sgmRNA produce N the vast majority of the time, and only occasionally ORF9b. /35
Because N's start codon precedes ORF9b's, ribosomes scanning their shared sgmRNA produce N the vast majority of the time, and only occasionally ORF9b. /35
But what if ORF9b had its own TRS occurring after the N start codon? In that case ORF9b would have its own sgmRNA, one that only produced ORF9b upon translation instead of mostly N. I think there's evidence for two such ORF9b-only TRS's. /36
The first overlaps with the ORF9b start codon. The core TRS—AATGGAC—is not a great match. Normally, I'd dismiss this, but in this case the extended homology is exceptional. (C is a good replacement for T, which is why I've included CCC in the extended homology) /37 

The second possible ORF9b-only TRS is a bit downstream & is also suboptimal—AAATGCAC. It overlaps with an in-frame start codon that would lead to a slightly truncated version of ORF9b. /38 

Notably, the near-universal C28311T improves this TRS's extended homology, & the likely advantageous C28312T—found in BQ.1.1 & synonymous in N—further enhances. The similar growth advantages of C28312T & C28313T are consistent w/the TRS hypothesis. 39/ 

Both ORF9b-only TRSs I'm proposing are suboptimal, meaning they'd be much less efficient at causing the RdRp to jump to the TRS-L & make a functional sgmRNA than an ideal TRS. But not having to share these TRS's with N may largely negate this disadvantage. /40
Example w/made-up numbers: From their shared sgmRNA, let's say N is translated 90% of the time & ORF9b 10%. And say the ORF9b-only TRS is 20x less efficient than an ideal TRS. The ORF9b-only TRS would increase ORF9b expression by 50%. /41
5) Alpha's 3-nuc N:D3L Mutation
Virtually all mutations involve a single nucleotide change. Two-nuc mutations are extremely rare & usually indicate strong selection pressure. If you see a 3-nuc mutation, you know something very unusual is going on. What's with Alpha's N:D3L? 42/
Virtually all mutations involve a single nucleotide change. Two-nuc mutations are extremely rare & usually indicate strong selection pressure. If you see a 3-nuc mutation, you know something very unusual is going on. What's with Alpha's N:D3L? 42/

N:D3L only makes sense as enhancing the suboptimal ORF9b-only TRS I discussed a few tweets up. It improves the core TRS and expands the already impressive extended homology on both ends, increasing production of ORF9b sgmRNA. /43 

This has been noted by the masterful @KroganLab. Alpha had the highest ORF9b sgRNA expression of any variant and, perhaps not coincidentally, remains the "the best overall innate immune suppressor" of any variant. 44/ https://t.co/NF0Iz55IrQnature.com/articles/s4158…


The TRS-enhancing nature of N:D3L was also noted by @Thushan_deSilva & @bioinfomatt in the excellent paper below which also provides a convincing explanation ("copy-choice recombination") of how this 3-nuc mutation occurred. /45nature.com/articles/s4200…
6) Delta & Alpha's ∆A28271
Another brief explanation is required for the next two on the list, which, in my view, seal the case for the importance of ORF9b. They involve something called a "Kozak sequence." 46/
Another brief explanation is required for the next two on the list, which, in my view, seal the case for the importance of ORF9b. They involve something called a "Kozak sequence." 46/
Some start codons (ATG) in an mRNA are more easily read by ribosomes due to the nucleotides that surround the them. The Kozak sequence describes the best/worst nucleotides in human cells at each position near a given start codon. 47/ 

In this diagram, the taller the letter, the more the corresponding nucleotide at that position favors start codon recognition by ribosomes. The -3, -1, and +4 positions have the largest effect, with -3 being the most influential. 48/ 

A bad Kozak sequence means a start codon is more likely to be passed over by the ribosome (leaky scanning). A bad Kozac sequence for N would reduce its translation, but it would *increase* ORF9b's translation. 49/ 

Above the Kozac chart is the Kozak sequence for N in wild-type (WT). At the most crucial position, -3, N has the best nuc (A), so it's Kozac sequence is pretty good. Now we come to one of the most remarkable instances of convergent evolution in the pandemic. /50 

Both Alpha *and* Delta had a 1-nucleotide deletion at this most important Kozac position, changing the -3 nuc from the best nuc (A) to the worst (T). The result is the single largest possible drop in Kozac favorability, at the expense of N protein expression. /51 

But N's loss is ORF9b's gain! One-nucleotide deletions are rare. Yet the two most widespread pre-Omicron VOC had the exact same 1-nuc deletion, the very one that would increase ORF9b expression the most. This cannot be a coincidence. /52
7) Omicron's A28271T
If any hardened walls of skepticism remain standing this one will obliterate them. *All Omicron lineages* have A28271T, causing the exact same A→T change at -3 & maximum possible loss in N Kozac favorability, to the benefit of ORF9b expression! /53
If any hardened walls of skepticism remain standing this one will obliterate them. *All Omicron lineages* have A28271T, causing the exact same A→T change at -3 & maximum possible loss in N Kozac favorability, to the benefit of ORF9b expression! /53

8) The I5T Advantage
Almost all the fastest lineages now have ORF9b:I5T, namely XBB.1.9.1, XBB.1.9.2, & the fastest of all, XBB.1.16. Maybe the only important difference between XBB.1.5 & XBB.1.9.(1/2) is ORF9b:I5T, so it seems clear the latter's faster growth is due to I5T. /54
Almost all the fastest lineages now have ORF9b:I5T, namely XBB.1.9.1, XBB.1.9.2, & the fastest of all, XBB.1.16. Maybe the only important difference between XBB.1.5 & XBB.1.9.(1/2) is ORF9b:I5T, so it seems clear the latter's faster growth is due to I5T. /54

Is there any way to independently test ORF9b:I5T's advantage? Let's look at ORF9b:I5T in Delta. Almost all Delta + ORF9b:I5T were in France, so the bottom graph, indicating a 26% weekly growth advantage, is likely the most accurate here. /55 

We can also look at BA.2* and BA.5* during their periods of dominance. (Outside those periods, random effects loom too large.) In both, we see a consistent 15% weekly growth advantage. There seems little doubt ORF9b:I5T is advantageous. /56 

9) ORF9b convergence in chronics
Final evidence of ORF9b's relevance comes from chronic-infection sequences, which I have spent many, many hours documenting. Mutations found frequently in chronic infections often, months/years later, become dominant in circulating lineages. /57
Final evidence of ORF9b's relevance comes from chronic-infection sequences, which I have spent many, many hours documenting. Mutations found frequently in chronic infections often, months/years later, become dominant in circulating lineages. /57
In unimportant genes, like ORF8 or ORF7b, there are no convergent mutations in chronics. In ORF9b, there are at least two: the aforementioned I5T and D89E, the latter found in BA.2.3.20. I think it's likely they confer immune-evasive benefits. /58
And with that, I rest my case for the importance of ORF9b. Crowded by N, ignored in phylogenetic trees, ORF9b is the Rodney Dangerfield of viral genes: It gets no respect. So let's give ORF9b its due going forward. 59/END
Huge thank you to all the labs & lab workers who do the sequencing that makes all this possible. And to @Chaoran_Chen_ for CovSpectrum, to @corneliusroemer & the @nextstrain crew, @theosanderson for GenSplore. This thread made extensive use of NextClade, GenSplore, & CovSpectrum.
And thanks to @PeacockFlu, @siamosolocani, @Asinickle1, @solidevidence, @nzm8qs, @JosetteSchoenma, @RajlabN, @PaulLMMA, @gianlucac1, @alchemytoday, and @wolfeagle1989 for all the productive discussions we've had on these things. I've learned from all of you.
@AltenbergLee If you meant how many nucleotides in the entire genome, it varies between branches, but most now have somewhere between 90-105 nucleotide substitutions relative to the original Wuhan strain. That's not counting deletions, of which there are several.
Excellent point here by @siamosolocani, who discovered that ORF9b mutations played an important role in the surprising success of BA.5.1 in Japan.
https://twitter.com/siamosolocani/status/1673662023740141571
• • •
Missing some Tweet in this thread? You can try to
force a refresh