1/ Thrilled to share our latest breakthrough in #CancerResearch! We've developed a novel machine learning model to predict patient response to immunotherapy. 🧬💊🔬 #Immunotherapy #AI. biorxiv.org/content/10.110…
2/ We analyzed eight cohorts of 2881 ICB-treated patients across 18 solid tumor types, the largest dataset to date, examining diverse clinical, pathologic, and genomic features. 

3/ We investigated 20 machine-learning models and developed a new pipeline to identify the most predictive model for ICB response. Our final model, named LORIS, leverages six commonly measured clinical features to predict ICB objective response using the RECIST criteria. 

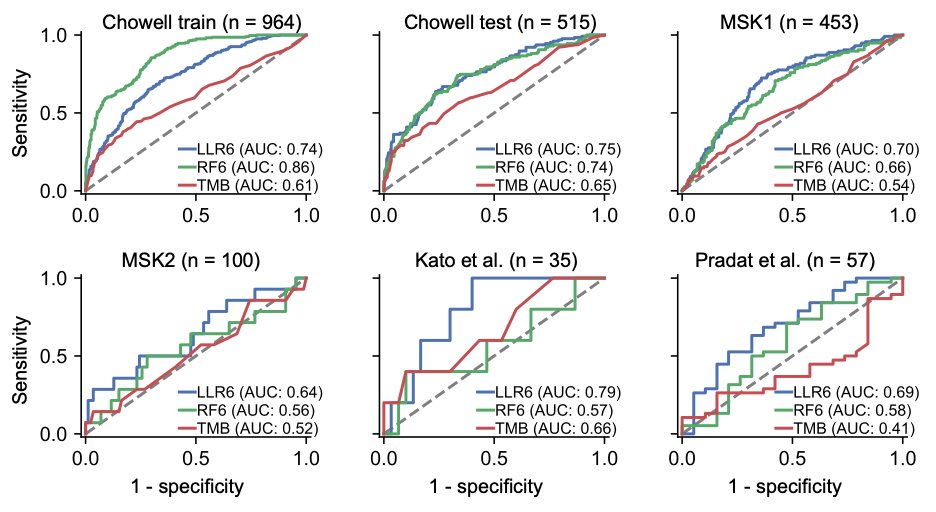
4/ Impressively, LORIS shows a negligible performance difference between training and test data, indicating robust generalizability and resistance to overfitting. This is a significant improvement over existing methods. #PrecisionMedicine #HealthTech 
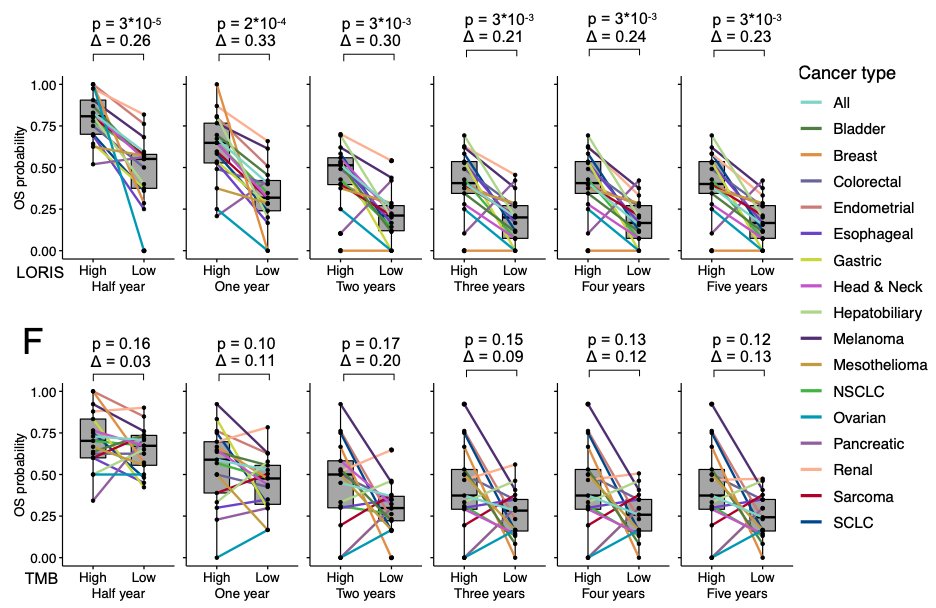
5/ LORIS goes beyond predicting patients’ response to immunotherapy. It also forecasts their short-term and long-term survival, both of which are critical clinical considerations. Notably, the TMB (Tumor Mutational Burden) biomarker lacks this predictive capacity. 
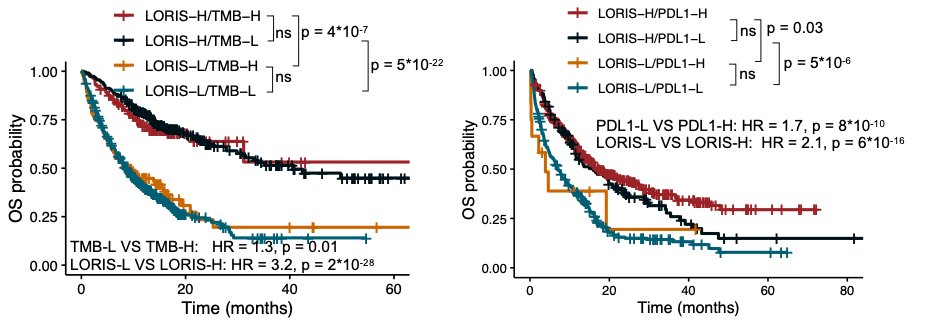
6/ One of the key findings is that LORIS can identify patients with low TMB or low PD-L1 (Programmed Death-Ligand 1) who can still benefit from immunotherapy. This could open up treatment options for many more patients. #CancerTreatment 
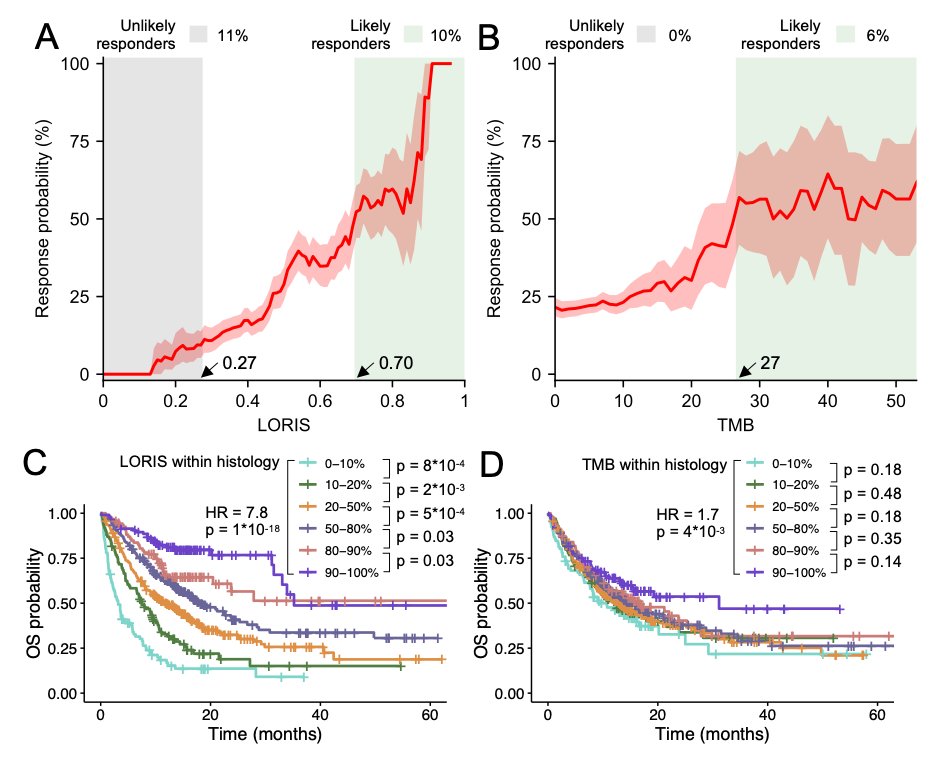
7/ Unlike many machine learning models, LORIS is transparent and interpretable, making it easier for clinicians to use.
8/ Most importantly, LORIS showcases a near-monotonic relationship with ICB response probability and patient survival, providing a consistent and accurate way to identify likely responders and exclude likely non-responders to immunotherapy. #MachineLearning #Healthcare 
9/ Using the same methodology, we've also developed a specific model for NSCLC (Non-Small Cell Lung Cancer), the cancer type with the largest sample size in our dataset. #LungCancer
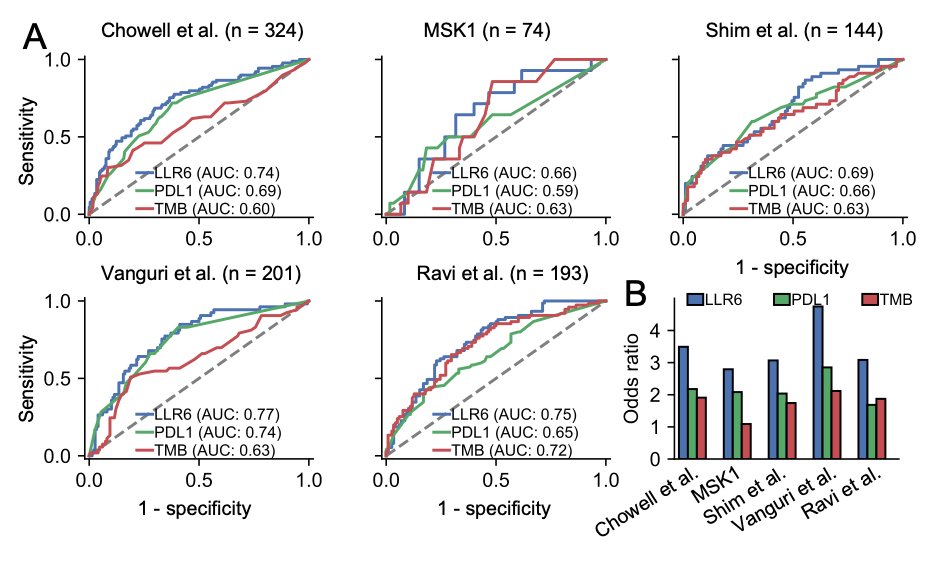
10/ This model also outperforms existing clinical biomarkers such as TMB and PD-L1, with an odds ratio for objective response ranging between 3 to 5, demonstrating the potential of our approach in modeling cancer-type-specific ICB response. #CancerCare 
11/ While our study is retrospective and more prospective studies are needed, the results are promising. As we continue to improve our understanding of tumor immunology, we hope to develop even more accurate models for personalized therapy. #FutureOfMedicine #CancerFight
12/ In summary, our study represents a significant step forward in the use of AI in cancer treatment. By accurately predicting patient response to immunotherapy, we can help more patients receive the most effective treatment for their specific cancer. #AIinHealthcare #EndCancer
13/ A huge shout-out to our co-authors @TiangenC, @learningbioinfo, @NCIEytanRuppin, @theNCI, and others. Also, a big thank you to our amazing collaborators @diegochowell @lucmorrisnyc and many more!!
@threadreaderapp unroll please
• • •
Missing some Tweet in this thread? You can try to
force a refresh

 Read on Twitter
Read on Twitter








