This paper from Linfa Wang’s group to which @tylernstarr & I contributed illustrates interesting points about
1⃣ Importance of imprinting for antibody specificity to SARS-like CoV
2⃣ How it may be easier to broadly neutralize animal sarbecoviruses than human #SARSCoV2 variants
1⃣ Importance of imprinting for antibody specificity to SARS-like CoV
2⃣ How it may be easier to broadly neutralize animal sarbecoviruses than human #SARSCoV2 variants
https://twitter.com/CheeWahTan2/status/1684344961700093953
Briefly, the study (which was led by Wan Ni Chia @CheeWahTan2 @LokShee @linfa_wang), isolated antibodies from person who had been infected by SARS-CoV-1 in ~2003, then received Pfizer SARS-CoV-2 vaccine in 2021.
So unlike most people in the world, whose first immunological exposure to a SARS-related CoV was to an early SARS-CoV-2 strain via either infection or vaccination in 2020 or 2021, this person’s immune system had been “imprinted” by SARS-CoV-1 prior to their COVID vaccine.
Three antibodies (E7, F1, and F5) effectively neutralized pseudoviruses with spikes from SARS-CoV-1 and SARS-CoV-2, as well as other animal SARS-related coronaviruses like WIV1 & GX-pangolin.
So these antibodies broadly neutralize many animal sarbecoviruses (SARS-related CoVs).
So these antibodies broadly neutralize many animal sarbecoviruses (SARS-related CoVs).

Our group mapped the functional epitope of these three antibodies (E7, F1, and F5) using deep mutational scanning, and showed all three had escape mutations at sites 408, 411, and 414. 

Our escape calculator (), which integrates deep mutational scanning from @yunlong_cao, shows sites 408, 411, & 414 are rarely targeted by neutralizing antibodies elicited by SARSCoV2 vaccines/infections in humans without prior SARS-like virus exposure. https://t.co/1zZU1XxXyhjbloomlab.github.io/SARS2-RBD-esca…


However, site 408 is a major focus of antibodies elicited by SARSCoV2 vaccination of humans with prior SARS-CoV-1 infection.
The differences between image shown here & post immediately above illustrate how profoundly immune imprinting shapes antibody specificity.
The differences between image shown here & post immediately above illustrate how profoundly immune imprinting shapes antibody specificity.

At first this seems exciting: imprinting with SARS-CoV-1 followed by boosting w SARS-CoV-2 can elicit antibodies that neutralize sarbecoviruses that only have ~76% spike protein identity!
But there are significant caveats when you dig a bit more deeply…
But there are significant caveats when you dig a bit more deeply…
The antibodies E7, F1, & F5 cross neutralize both SARS-CoV-2 & animal sarbecoviruses w spike identities of only ~76%, but they have greatly reduced neutralization against newer SARS-CoV-2 variants (like XBB) even though XBB has >96% spike amino-acid identity to early SARS-CoV-2. 
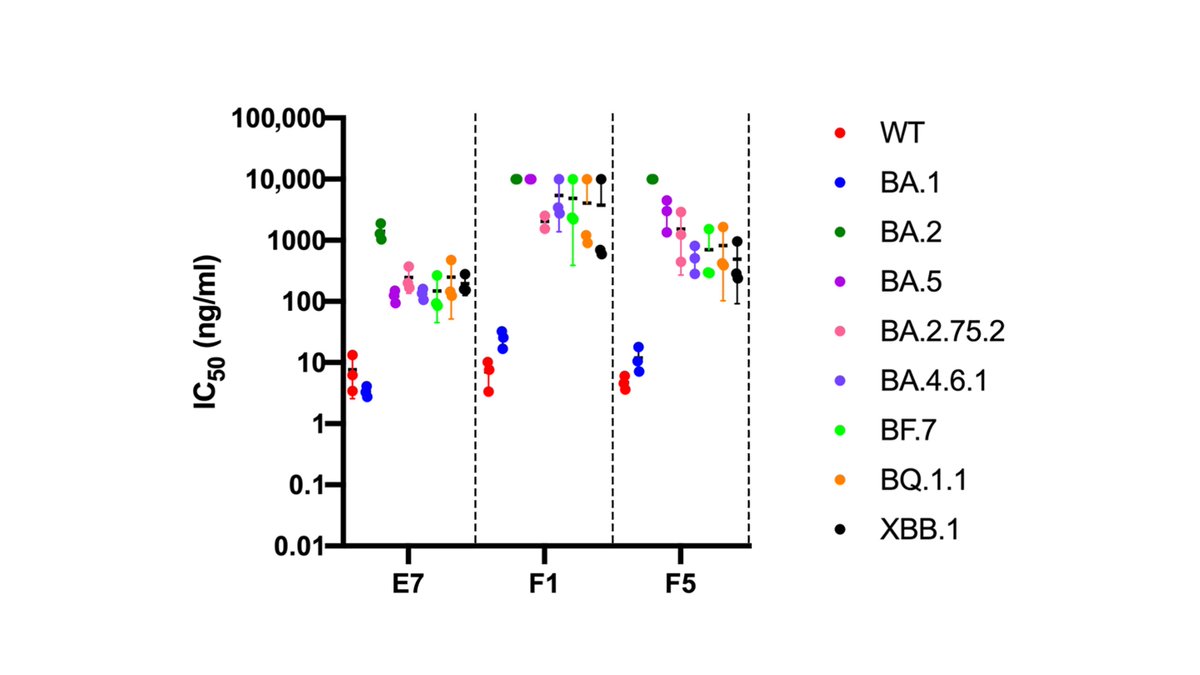
This underscores a point that is insufficiently appreciated in discussions of pan-sarbecovirus vaccines & antibodies:
It is probably EASIER to cross-neutralize animal sarbecoviruses w ~75% spike identity than human SARS-CoV-2 variants w >95% spike identity.
It is probably EASIER to cross-neutralize animal sarbecoviruses w ~75% spike identity than human SARS-CoV-2 variants w >95% spike identity.
This observation may sound counterintuitive, but it has a good explanation: relative rates of genetic evolution (how much the sequence changes) and antigenic evolution (extent of antibody escape) can depend on the host species.
Species-specific differences in antigenic evolution are best characterized for influenza virus, where similar viruses undergo much more rapid antigenic evolution in humans than in pigs (eg, )microbiologyresearch.org/content/journa…
Reason is easy to understand:
Humans are long-lived, so viruses get a big advantage from antibody-escape mutations that let them re-infect previously exposed individuals.
This is a lot less true for pigs, where a large fraction of population is always naïve.
Humans are long-lived, so viruses get a big advantage from antibody-escape mutations that let them re-infect previously exposed individuals.
This is a lot less true for pigs, where a large fraction of population is always naïve.

Although ecology of SARS-like CoVs less understood, probably antigenic mutations confer less advantage in bats than humans for similar reason.
Indeed, another study Linfa Wang shows less antigenic evolution in animal vs human SARS-like viruses: nature.com/articles/s4156…
Indeed, another study Linfa Wang shows less antigenic evolution in animal vs human SARS-like viruses: nature.com/articles/s4156…
Taken together, this illustrates how exposure to diverse animal SARS-related CoVs can elicit antibodies that broadly neutralize such viruses.
But this may be of only modest use against SARS-CoV-2 variants that evolved in humans under pressure to escape neutralizing antibodies.
But this may be of only modest use against SARS-CoV-2 variants that evolved in humans under pressure to escape neutralizing antibodies.
• • •
Missing some Tweet in this thread? You can try to
force a refresh

 Read on Twitter
Read on Twitter