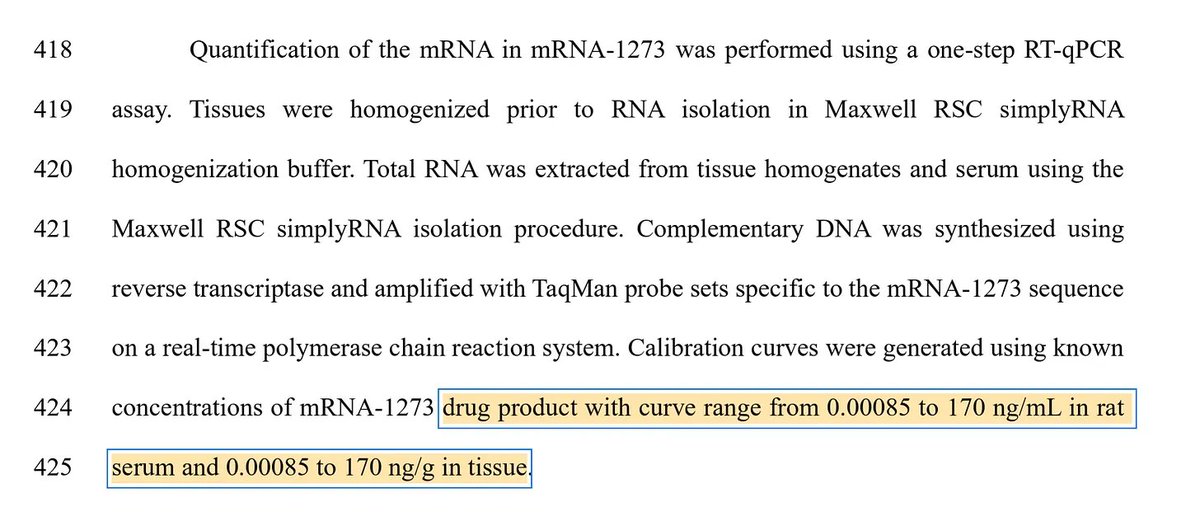
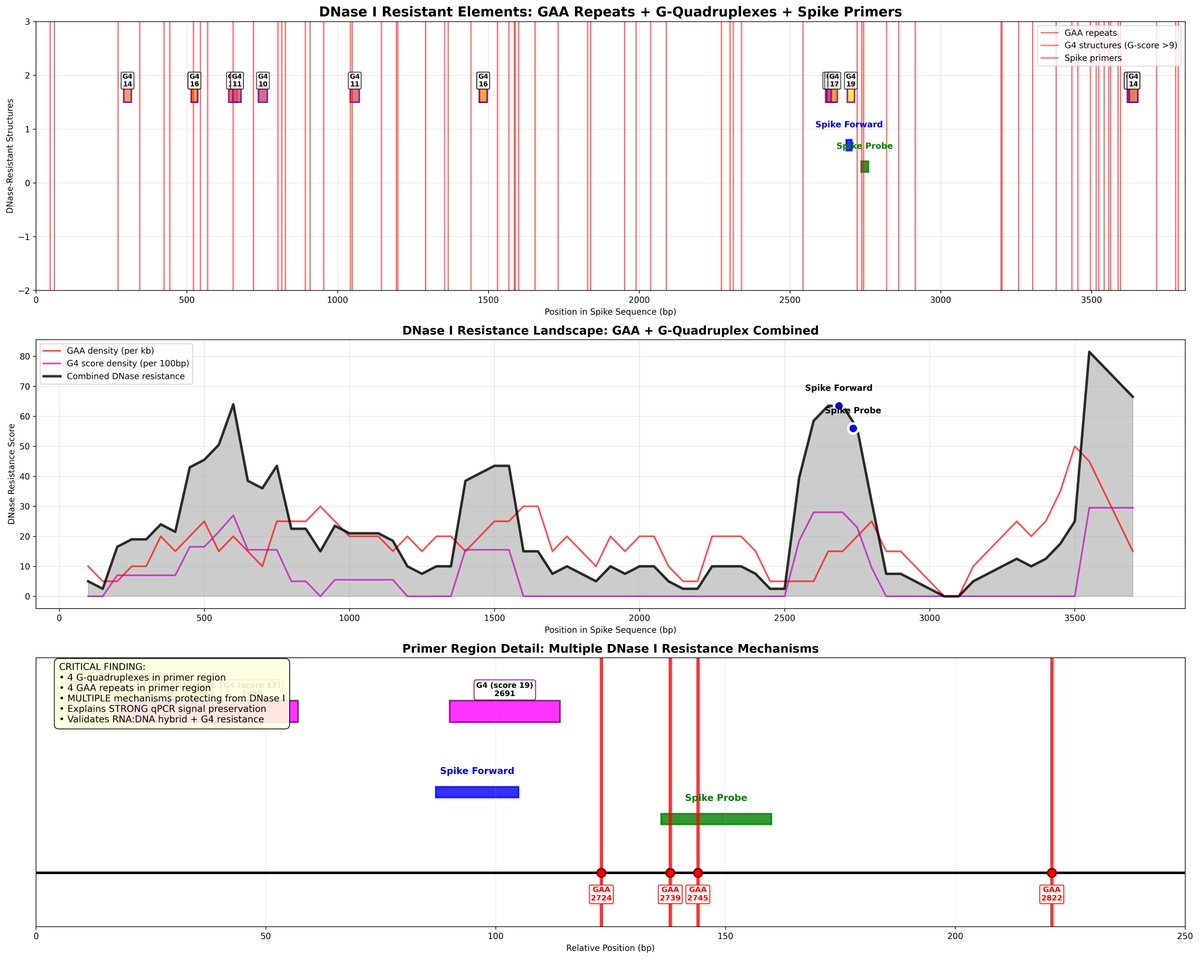
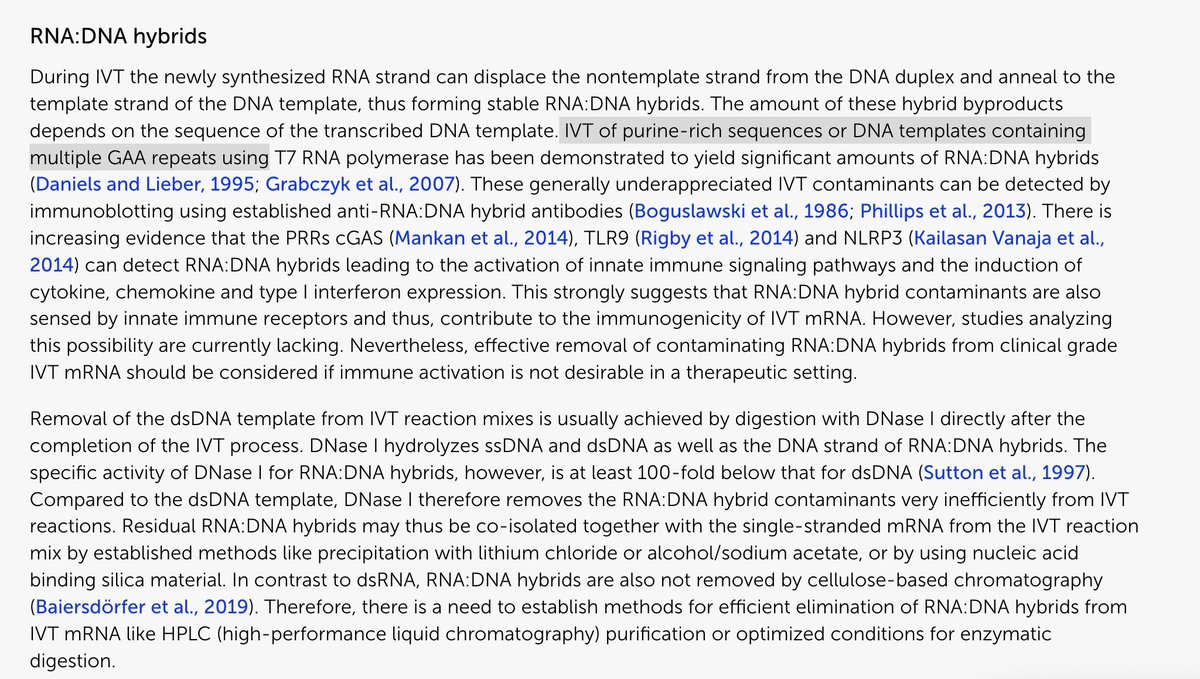
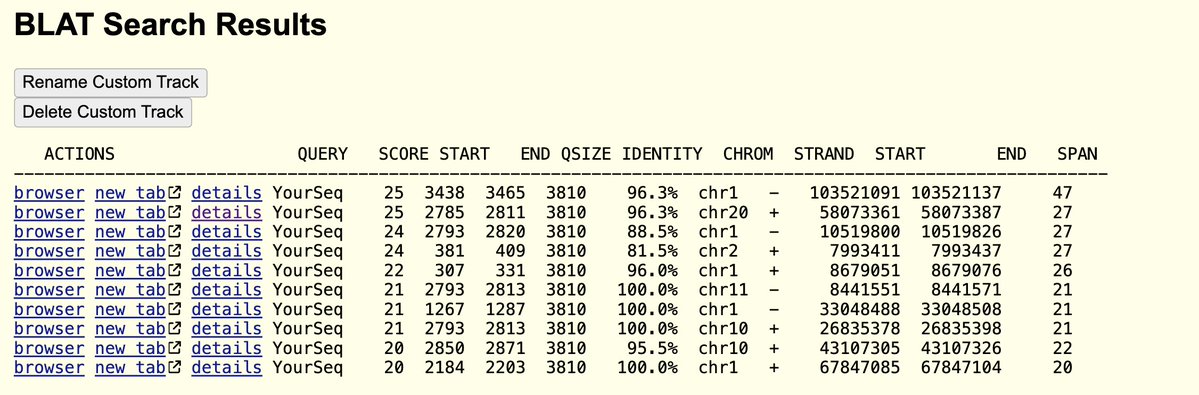
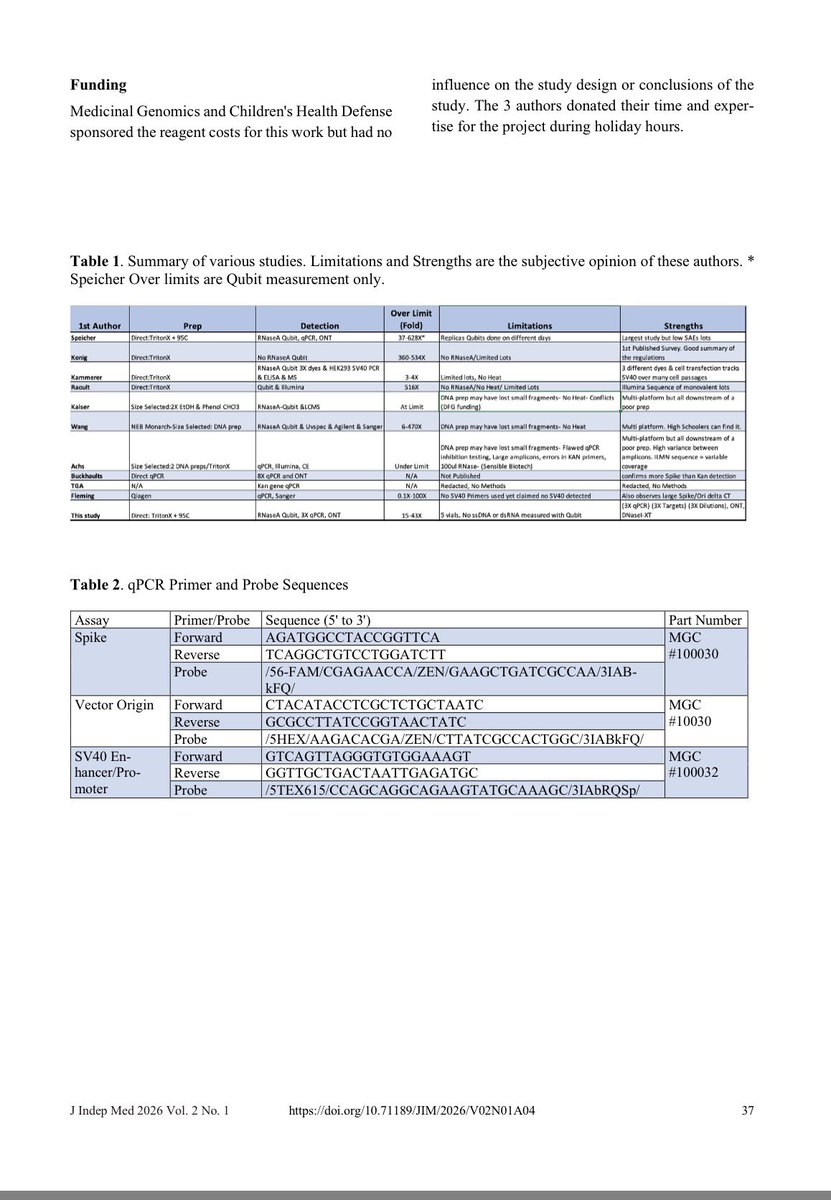
The plasmid they used in the above study doesn’t have an SV40 72 base enhancer element that Dean et al demonstrate to enhance these efficiencies due to its nuclear targeting sequence. 

They cut the vector at BsaI leaving that red arrow.
They other 25-40% of the vector has no GFP tracer for integration. That will under estimate the linear risk if they normalized based on molecules.
The vector also has this in it.
Not a NTR
pubmed.ncbi.nlm.nih.gov/17597793/
They other 25-40% of the vector has no GFP tracer for integration. That will under estimate the linear risk if they normalized based on molecules.
The vector also has this in it.
Not a NTR
pubmed.ncbi.nlm.nih.gov/17597793/
• • •
Missing some Tweet in this thread? You can try to
force a refresh