🧵T cell VDJ Recombination:
Here we will look more into the T cell receptor creation.
Here we will look more into the T cell receptor creation.
1/ The T cell will begin the arrangement of its Beta chain of the T cell receptor. This works in a competitive environment as both sets of genes from mom and dad compete to see which one is able to rearrange a successful Beta chain first.
2/ The genes for the Beta chain are located on chromosome 7. They are divided into 3 categories of the Variable genes, the Diversity genes and the Joining genes. 

3/ There are about 60 different variable genes to choose from, usually represented as V1 to V60. There are 27 Diversity genes to choose from, represented D1 to D27. Then there are 6 different Joining genes to choose from represented as J1 to J6.
4/ The enzymes that regulate the recombination process will randomly choose 1 Variable, 1 Diversity and 1 Joining gene.
5/ There are many possible combinations of these genes. During this process all the genes between the selected segments are removed and broken down. It starts by picking a single D and J segment and putting them together and removing the DNA in between.
6/ Then it continues to take a random V gene and bind it to the already connected DJ segment removing the DNA between them. This brings together a single VD&J segment to make a Beta chain which will be transcribed. 

7/ This leaves the T cells with just 1 choice for each of these VDJ genes. Not every gene segment has to be removed in recombination. For example, if the RAG enzymes select V20 out of 60 possible gene choices, all the genes before that are left in the DNA from V1 to V19.
8/ The same happens with the J genes. The genes after the one selected are left inside the DNA after the recombination. There are never any D segments left except the one chosen as all the rest are removed between them and the V and D segments.
9/ The first Beta chain to reach a successful arrangement and display it on the cell surface will win. The other genes for the opposite chromosome and the Delta/Gamma genes will all be inactivated.
10/ This is the process of allelic exclusion to ensure each T cell only makes 1 receptor. If the T cell fails to make a successful Beta chain for any reason with either allele, it will try to make a Gamma chain from both alleles of that gene.
11/ This acts like a rescue for the cell to try another receptor. About 95% of the T cells will have an Alpha/Beta receptor and 5% will end up with Delta/Gamma receptors.
12/ The next step will be to rearrange the alpha genes which are on chromosome 14. The alpha chain has only 2 gene categories to draw from. They are the Variable and Joining genes. There are about 70 possible genes in the Variable category to choose from, represented V1 to V70. 

13/ There are another 61 or so genes in the Joining Category to choose from, represented J1 to J61. The alpha genes have no Diversity category. The location for the Delta genes are actually inside the Alpha genes located between the Variable and Joining genes.
12/ A successful arrangement of the alpha chain removes the Delta genes so no possible arrangement of those genes can occur. When the T cell receptor forms it will create there loops called the Complementarity Determining Regions (CDR) loops. 
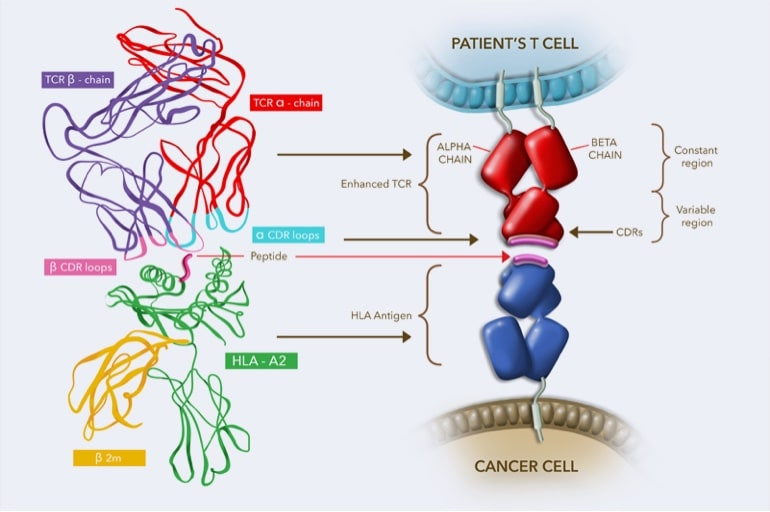
13/ The Variable gene is about 100 nucleotide long and makes up the CDR1 and CDR2 loops. The combination of the end of the V the D and the J all come together to make up the CDR3 loop. This makes CDR3 the hypervariable loop.
14/ Due to the fact that when the enzymes paste together the V, D, and J segments, some random nucleotides will be added or even removed. This creates vast diversity in this region of the TCR. 

15/ This happens with the Beta chain in big way as it has 3 genes coming together in this CDR3 region of the receptor. The alpha chain has CDR3 region made up of the connection of the V and J region. Its not quite and hypervariable as the beta chain.
16/ The CDR1 and CDR2 mostly interact with MHC which is why they need to be consistent. The CDR3 is what recognized the antigen peptide which is why it is hypervariable. This allow for a vast repertoire of T cells to be created.
• • •
Missing some Tweet in this thread? You can try to
force a refresh

 Read on Twitter
Read on Twitter











