Updates on several concerning variants, JN.1 (BA.2.86+L455S), JD.1.1(FLip+A475V), HV.1(EG.5+L452R).
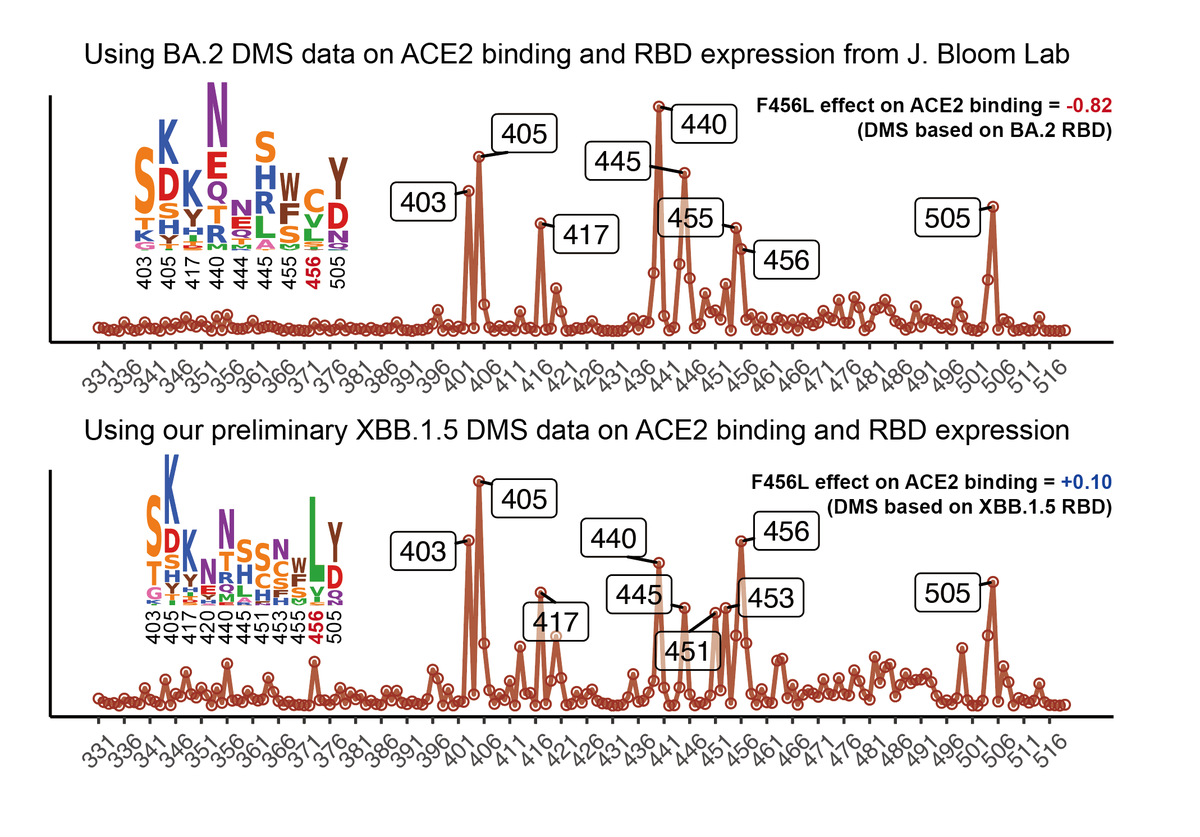
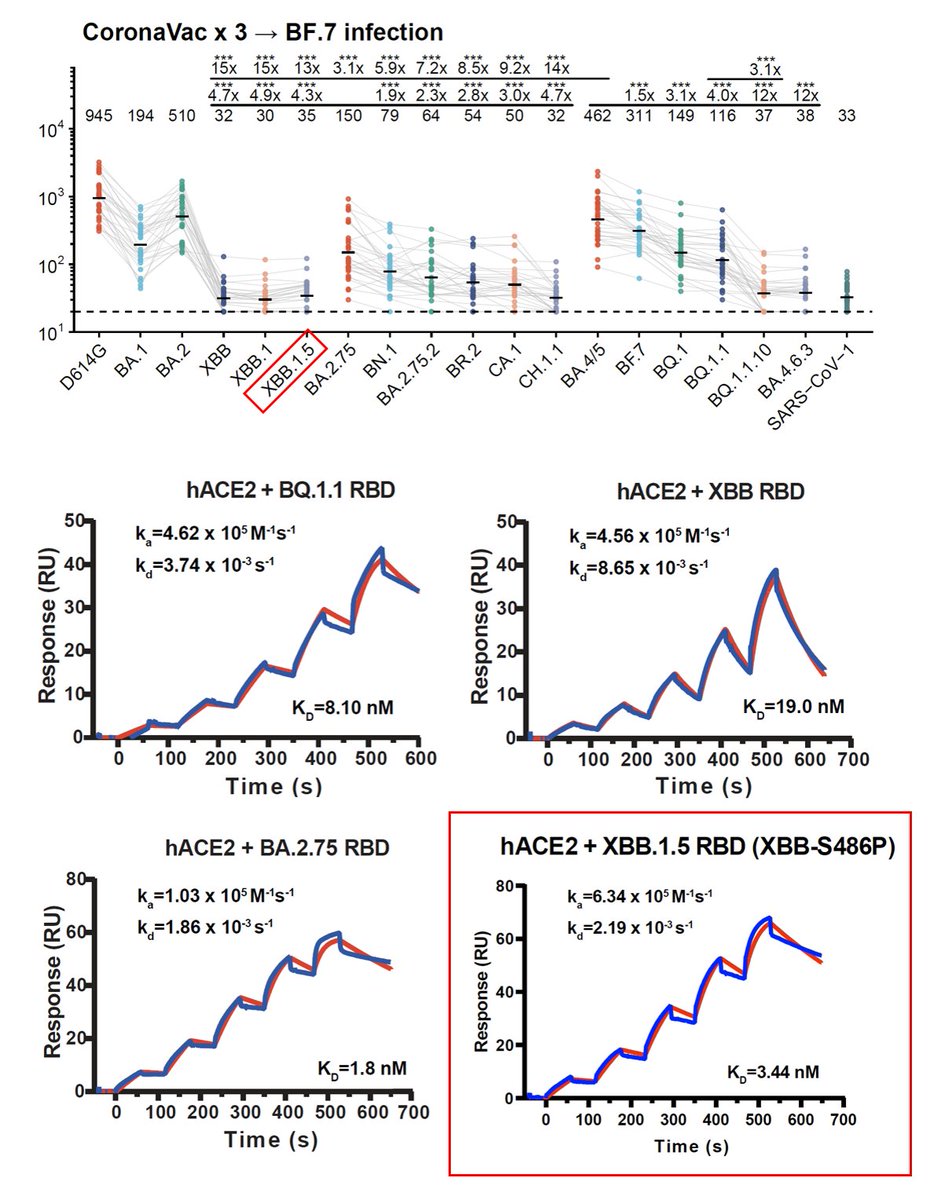
1) L455S on BA.2.86 (JN.1) greatly increases antibody evasion at the cost of ACE2 binding.
2) HV.1 and JD.1.1 are more evasive than FLip but display lower ACE2 binding as well. 1/3
1) L455S on BA.2.86 (JN.1) greatly increases antibody evasion at the cost of ACE2 binding.
2) HV.1 and JD.1.1 are more evasive than FLip but display lower ACE2 binding as well. 1/3

L455S mainly escapes Class 1 neutralizing antibodies, which made up for the weakness of BA.2.86 (vulnerable to Class 1 Abs). Of note, FLip + A475V could evade almost all of Class 1 Abs, which explains why we have seen so many A475V mutations on FLip variants recently. 2/3 

These data again emphasize that high ACE2 binding affinities, such as FLip (455F+456L) variants and BA.2.86, would allow fast collections of mutations that can further boost immune evasion. 3/3 

• • •
Missing some Tweet in this thread? You can try to
force a refresh

 Read on Twitter
Read on Twitter