What’s new in the world of SARS-CoV-2 accessory proteins? In this 🧵, I’ll recap their evolution so far & describe a possible new trend: partial or total deletion of ORF7a, ORF7b, & ORF8—particularly in GW.5.1.1, an XBB.1.19 + FLip lineage—plus an ORF9b/N bonus TRS surprise! 1/45



The SARS-CoV-2 accessory proteins are ORF3a, ORF6, ORF7a, ORF8, ORF9b, and N* (the last absent from Delta). While the non-structural & structural proteins are absolutely essential for replication, accessory proteins are not. 2/45
https://twitter.com/LongDesertTrain/status/1666220308599037955
In cell culture & in mice, deleting each accessory protein (one at a time) causes only slight-to-moderate reductions in viral replication & pathogenesis. ORF3a & ORF6 seem more crucial than ORF7a, ORF7b, and ORF8. (ORF9b & N* are excluded from these studies). 3/45 

Other studies have shown accessory proteins play important roles, often by mucking up normal cellular functions, especially innate immune defenses, like the interferon response. Some of the best work in this area has been by @KroganLab. 4/45
https://twitter.com/KroganLab/status/1704877701323178057
For a detailed look at some of the posited mechanisms by which accessory proteins evade innate immune responses, see the excellent, definitive review on this subject by @JudyMinkoff & @virusninja. 5/45
https://twitter.com/virusninja/status/1613229938177368075
Earlier this year, @PeacockFlu, @siamosolocani, & I wrote a paper describing the loss of ORF8 in globally widespread lineages, first in Alpha, then BA.5, XBB.1*, and XBC.1 (a Delta-Omicron recombinant). Below is a thread I wrote on ORF8’s demise. 6/45
https://twitter.com/LongDesertTrain/status/1645223059698139142
Eminent phylogenetics guru and genius-award winning @trvrb recently released a paper with @pavitrarc & @wcassias showing, through multiple lines of evidence, that ORF8 knockout has been positively selected for in SARS-CoV-2. 7/45
https://twitter.com/wcassias/status/1707460019061719289
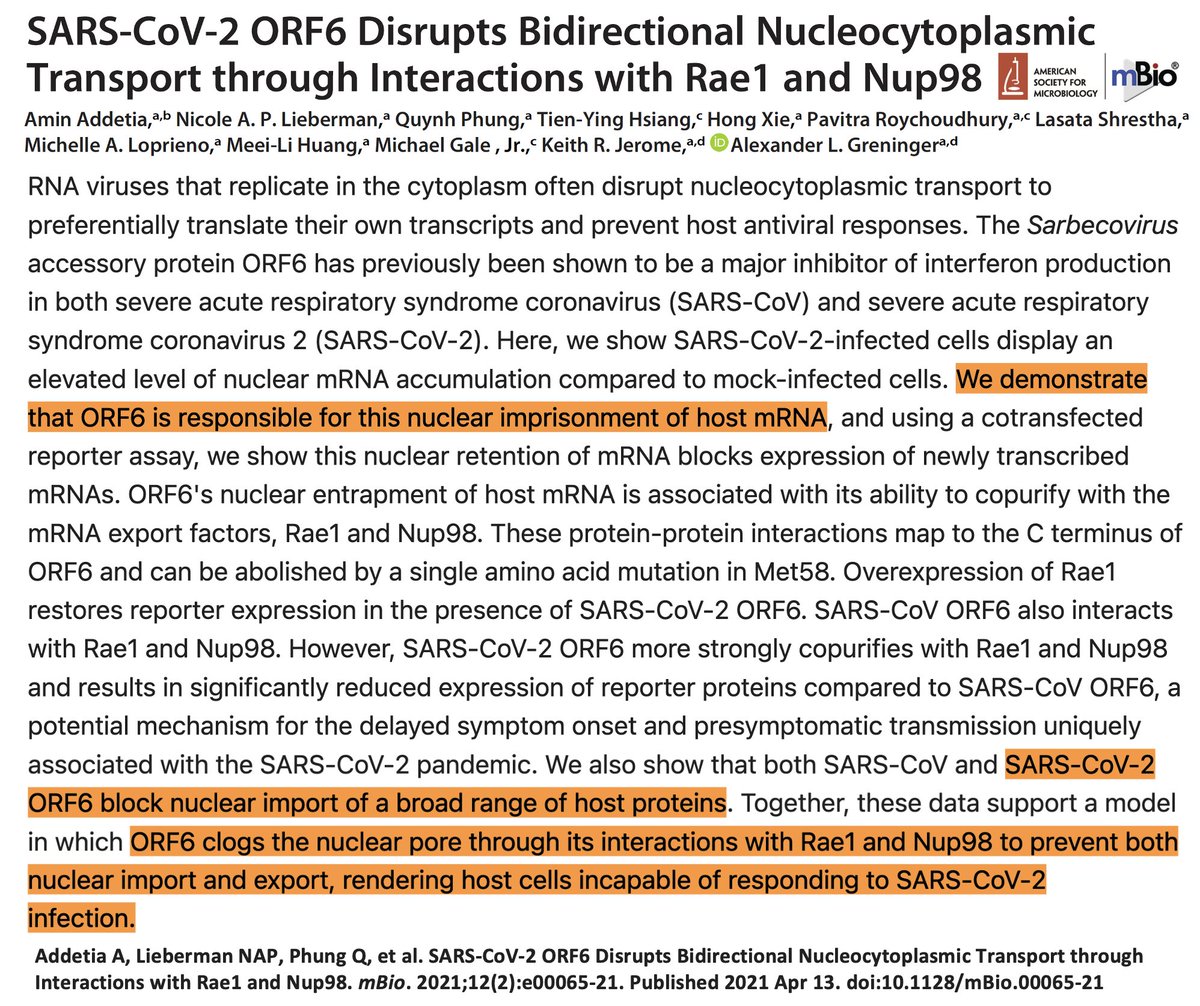
ORF3a has remained relatively untouched by large deletions, stop codons, and TRS destruction. So has ORF6, which @GreningerLab, @AminAddetia, & others have shown blocks import & export of cellular cargo, stonewalling our antiviral interferon response. 8/45 
ORF7a & ORF7b, on the other hand, have frequently been erased by large deletions, frameshift-causing deletions, & stop codons. In the thread below, I detail some evidence that there seems to be evolutionary pressure to reduce ORF7a expression. 9/45
https://twitter.com/LongDesertTrain/status/1652082424199643139
In that thread, I showed that C27389T, which virtually abolishes ORF7a expression, showed marked growth within each variant wave. But you can see that two variants did not fit the pattern: BA.2 and XBB. I think there is a reason for this—ORF6:D61L. 10/45
https://twitter.com/LongDesertTrain/status/1652082430650490882
ORF6:D61L is a rare bird—a 3-nuc mutation. It is present in all BA.2-derived lineages, including all XBB. It was also in BA.4 but not BA.5, & this may be one of the primary reasons BA.5 so easily outgrew BA.4—despite having an identical spike. 11/45 
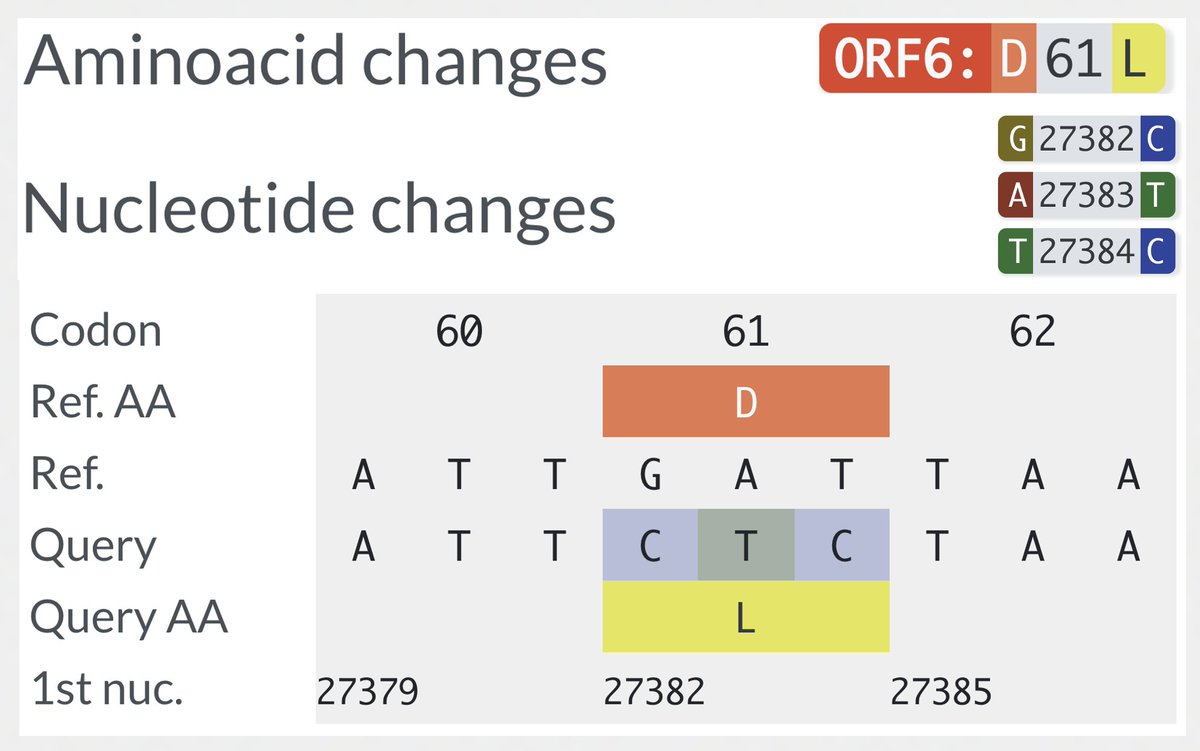
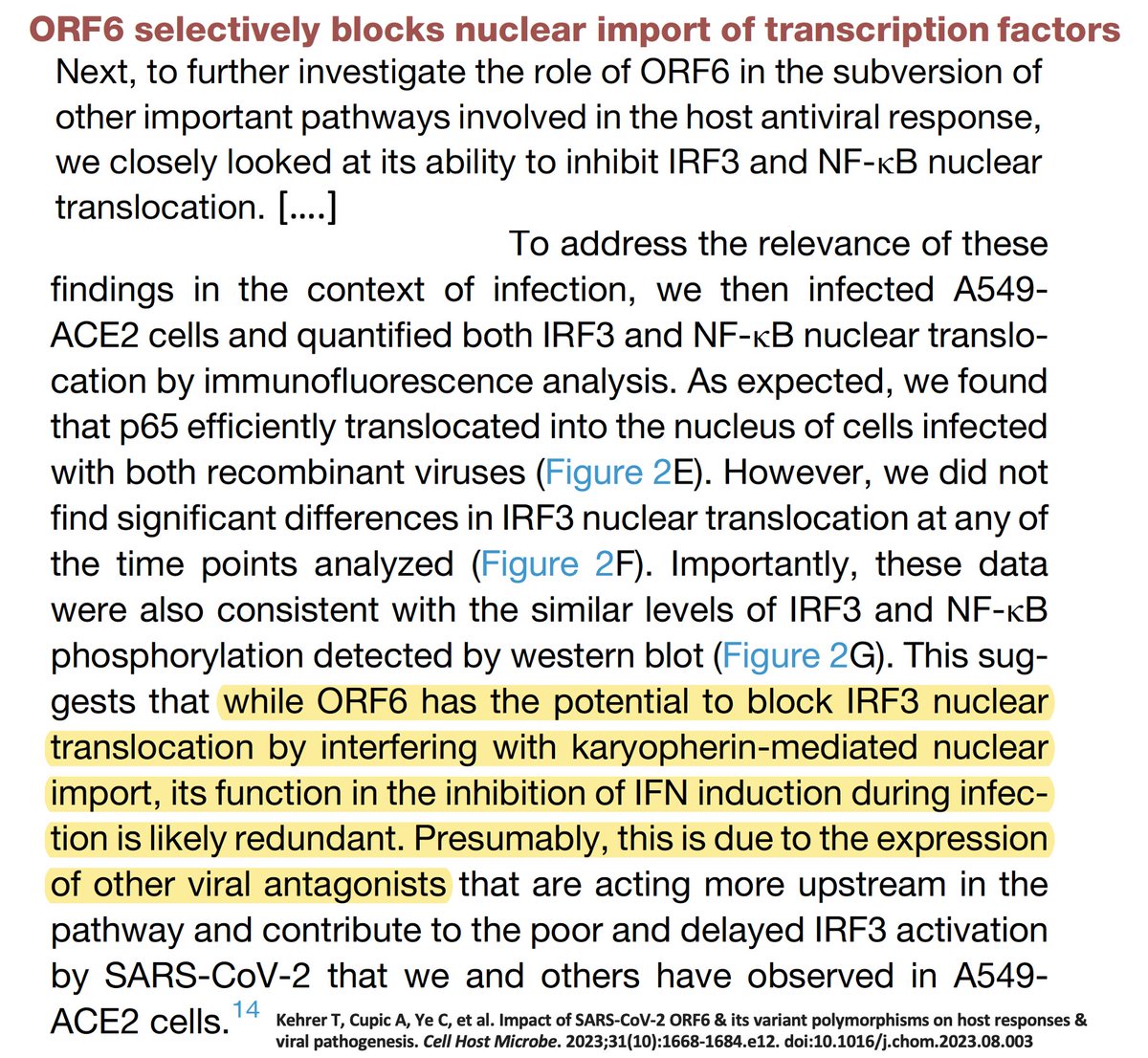
In a superb ORF6 paper published last month, @LisaMiorin & @KroganLab show that ORF6:D61L seriously impairs ORF6’s immune evasion function by reducing its ability to block cellular trafficking in & out of the nucleus. 12/45



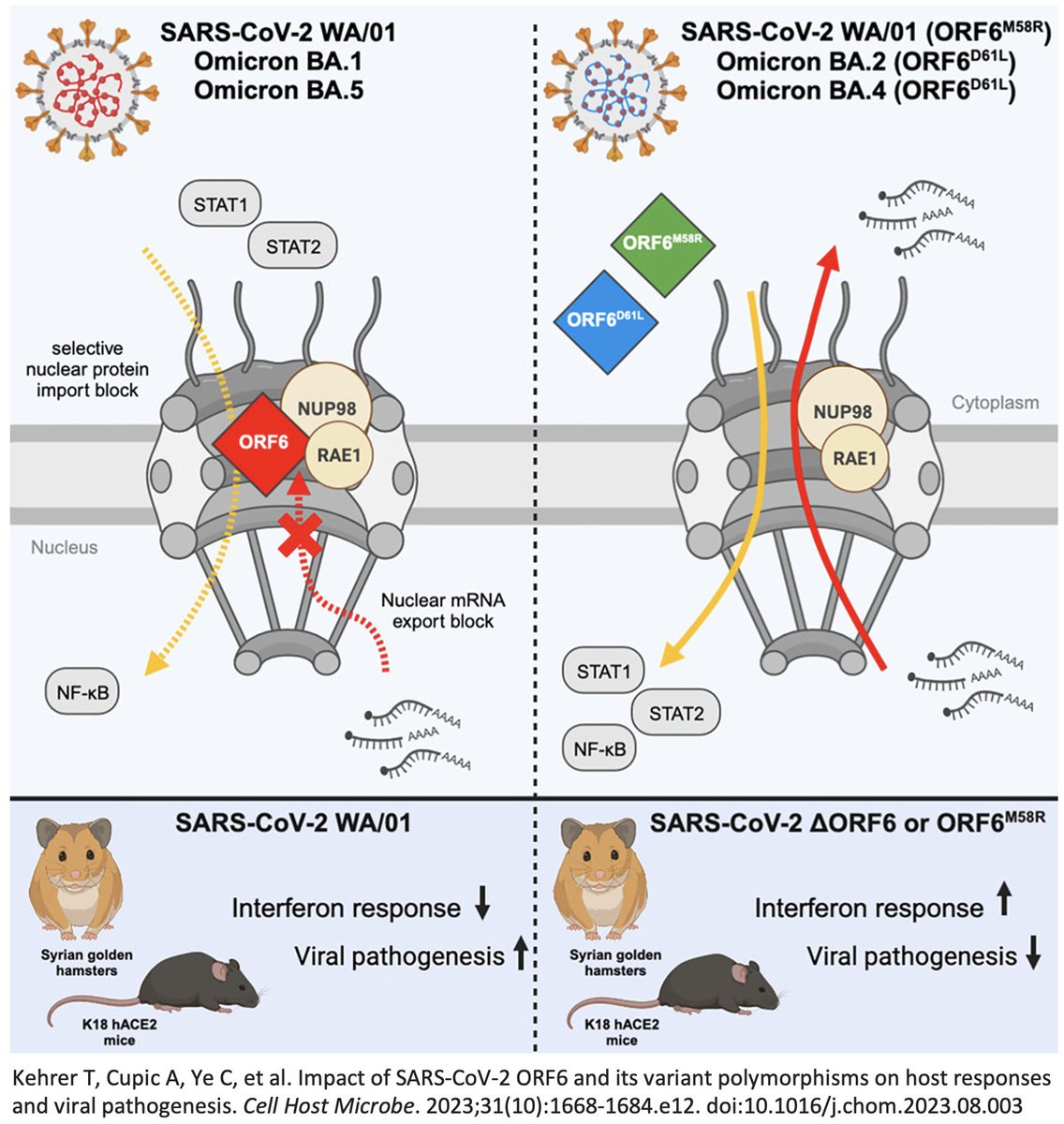
If D61L harms ORF6 function, why is it so prevalent? It may have just piggybacked on BA.2, which had a bevy of other beneficial mutations. And while a harmful 1-nuc mutation can revert easily enough, it’s much more difficult to walk back a 3-nuc mutation. 13/45
ORF6:D61L is a TRS-related mutation that almost certainly arose due to a “copy-choice” recombination event. D61L creates extended homology for the ORF7a TRS, which should increase its expression (i.e. create more copies of ORF7a mRNA). 14/45 

I use the term “TRS” a lot in this thread, & I know it’s confusing. That’s why I spent many hours putting together an explanation of what a TRS is and how it works. You can read it in tweets 4-21 in the thread below (if you haven’t already). 15/45
https://twitter.com/LongDesertTrain/status/1673488099014504449
A few functions of these accessory proteins are understood, but like most things in biology, most of what they do is unknown. But whatever their exact functions, I think there’s likely a large amount of overlap, esp. between ORF6 & ORF7ab—as @KroganLab has hinted at. 16/45 
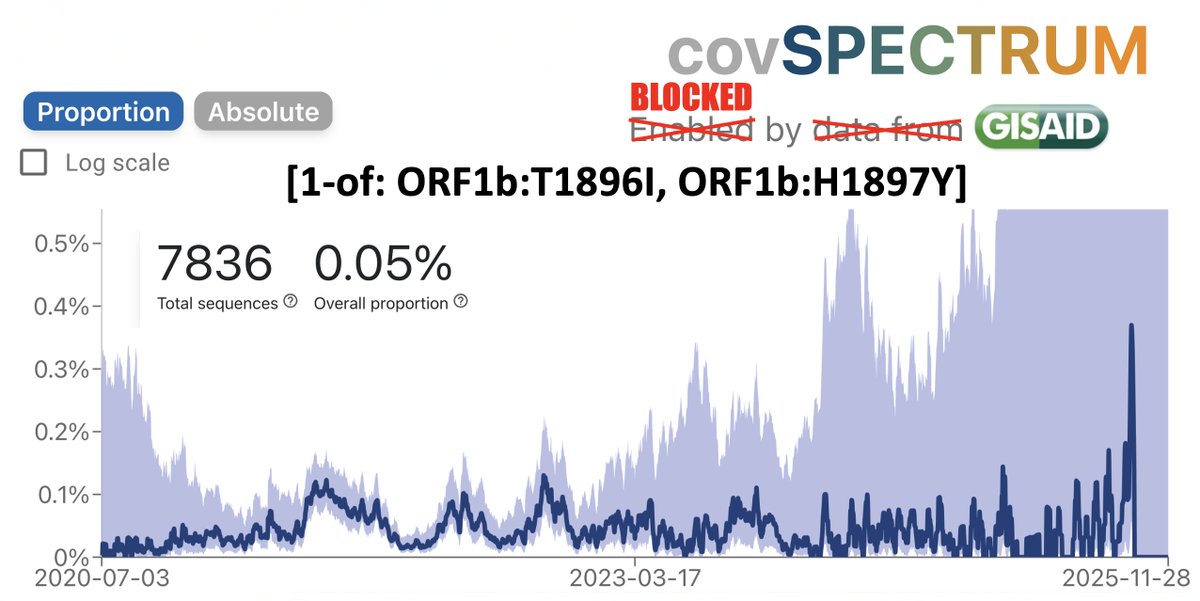
Witness the remarkable inverse relationship between the prevalence of ORF6:D61L (which hobbles ORF6) compared to the combined prevalence of three ORF7a TRS-killing mutations & large ORF7a deletions. (Y-axis scales are different to make the relationship clear). 17/45 
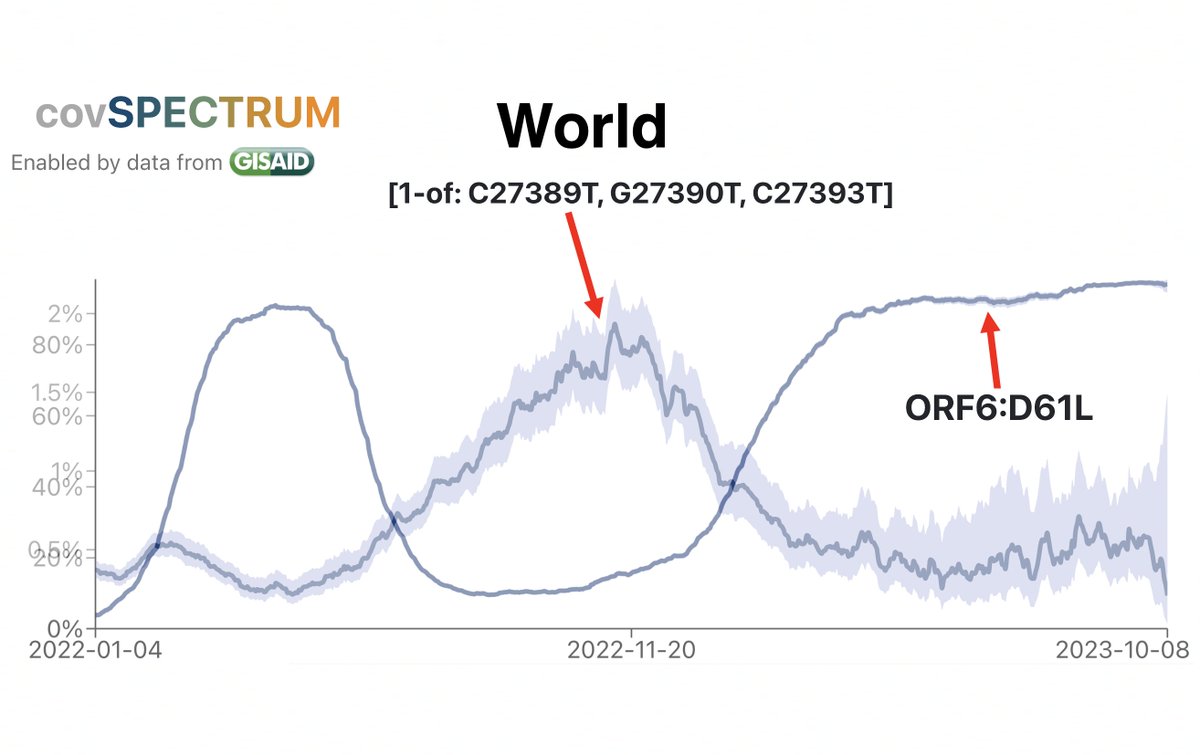
Due to the frequency of low-quality sequences, it’s much more difficult to accurately determine the prevalence of deletions. But a Cov-Spectrum search for deletions in ORF7a of about 25 or more nucleotides reveals a similar inverse trend with ORF6:D61L. 18/45 
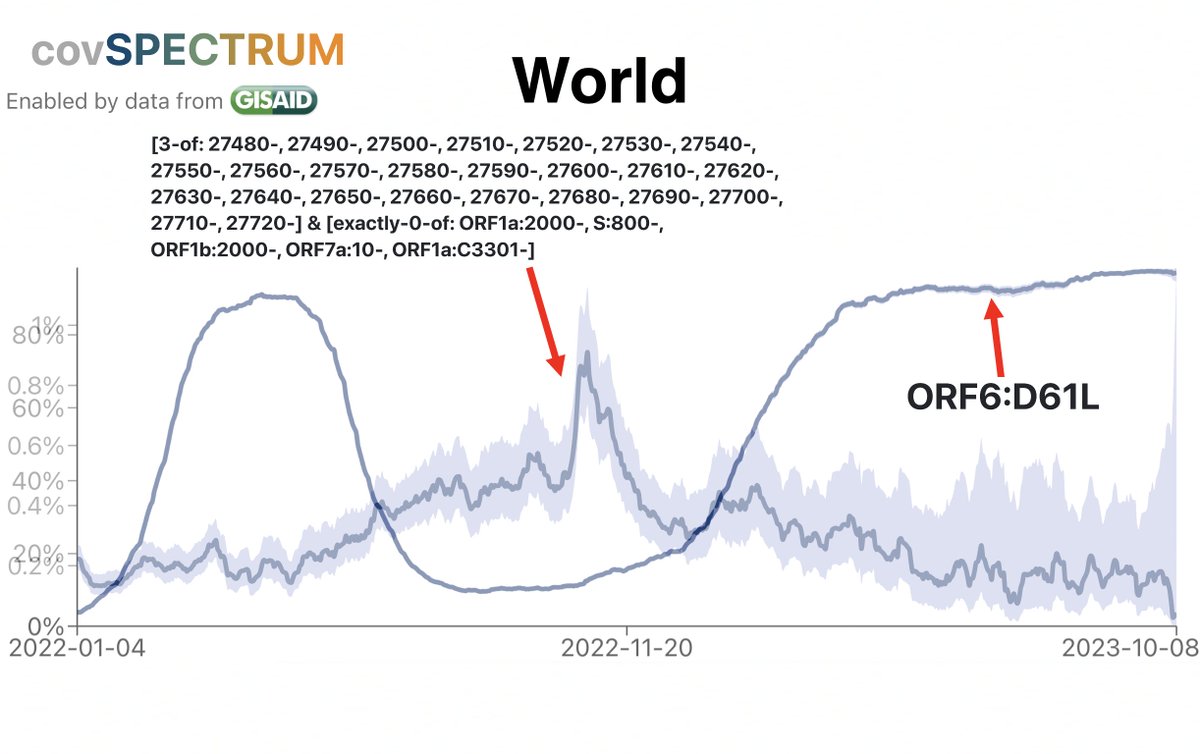
For reasons unknown, global trends in ORF7a ablation are enormously magnified in South Africa, with peaks in ORF7a-TRS-killers & large deletions ~20-fold higher—note identical y-axis scales. Any ideas, @Tuliodna, @Rajeev_the_king, @sigallab, @houzhou, @DarrenM98230782? 19/45 
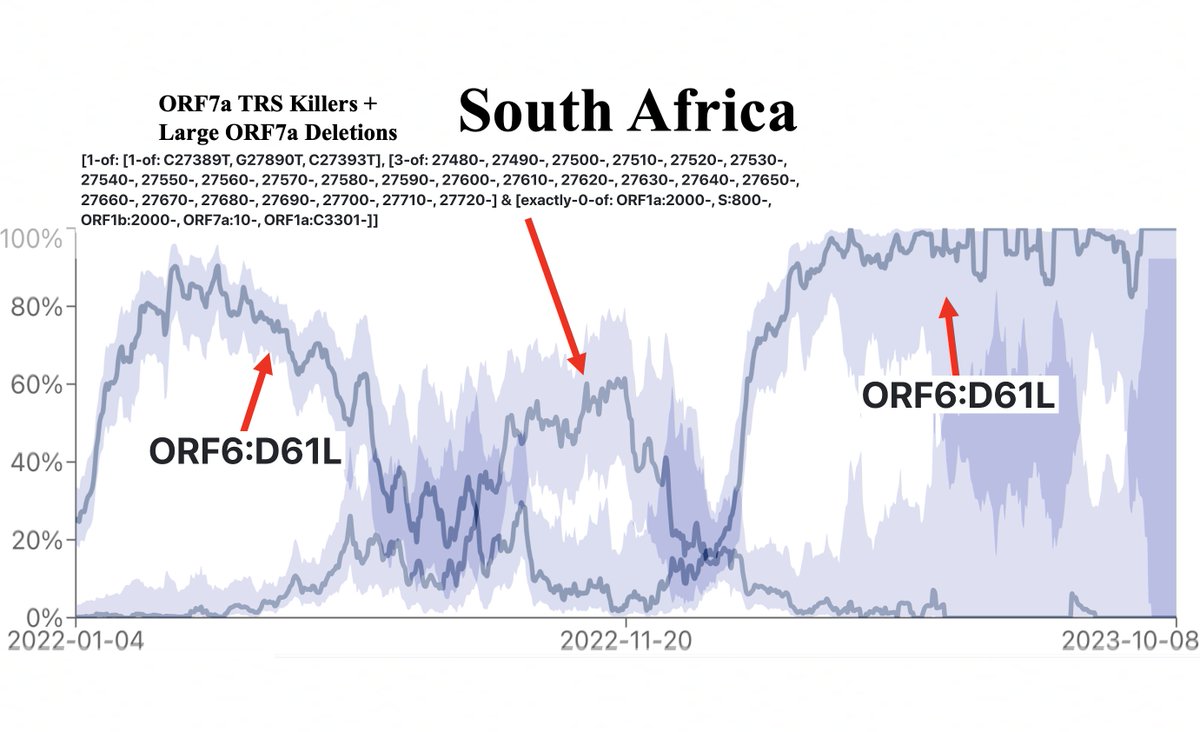
It seems that as long as ORF6 is fully functional and not hobbled by ORF6:D61L, ORF7a is disposable. But when ORF6:D61L is present, there is strong selection against mutations that knock out ORF7a, suggesting they may have overlapping functions. 20/45
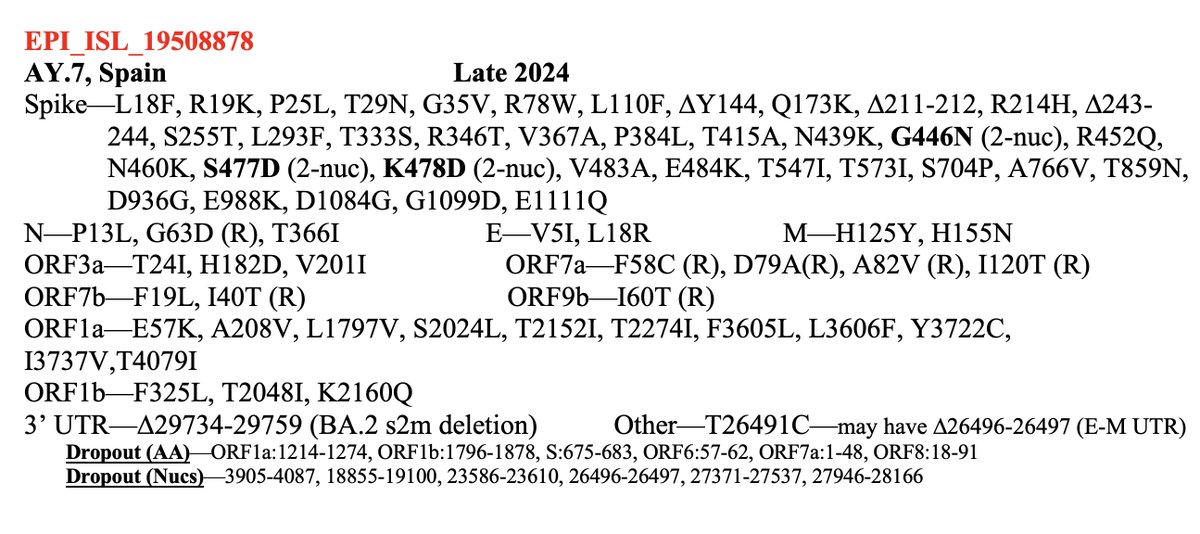
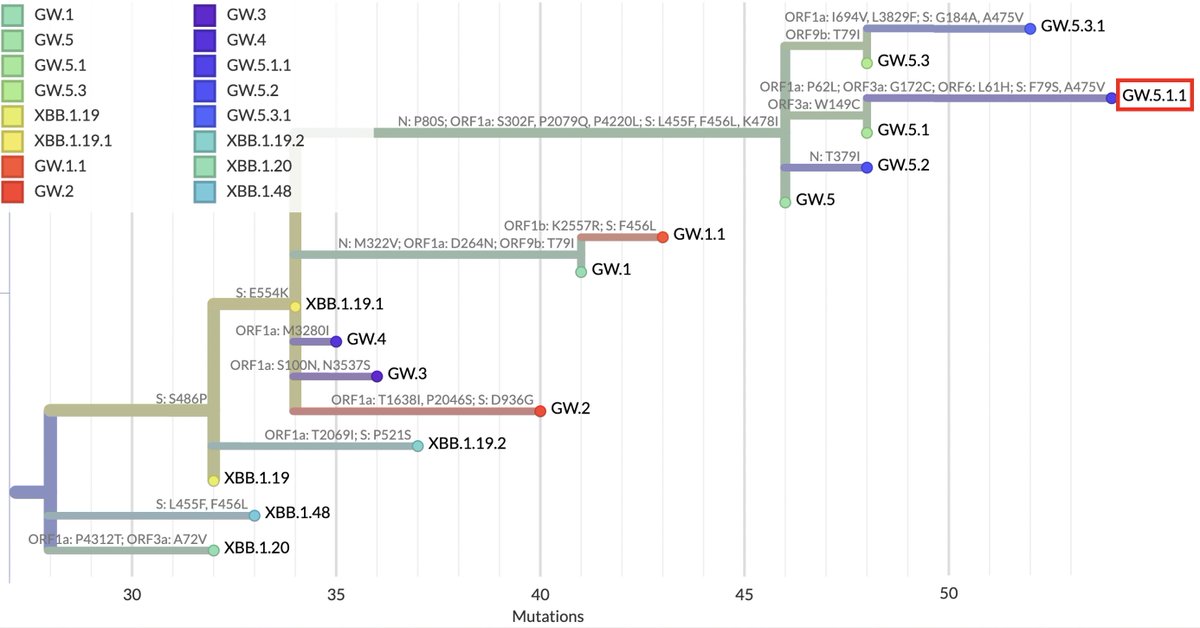
This may be relevant to the primary subject of this thread: the possible disappearance of ORF7a, ORF7b, and ORF8 from one of the fastest-growing SARS-CoV-2 lineages, GW.5.1.1—a rare XBB.1* that had an intact ORF8—and similar deletions in other lineages. 21/45 
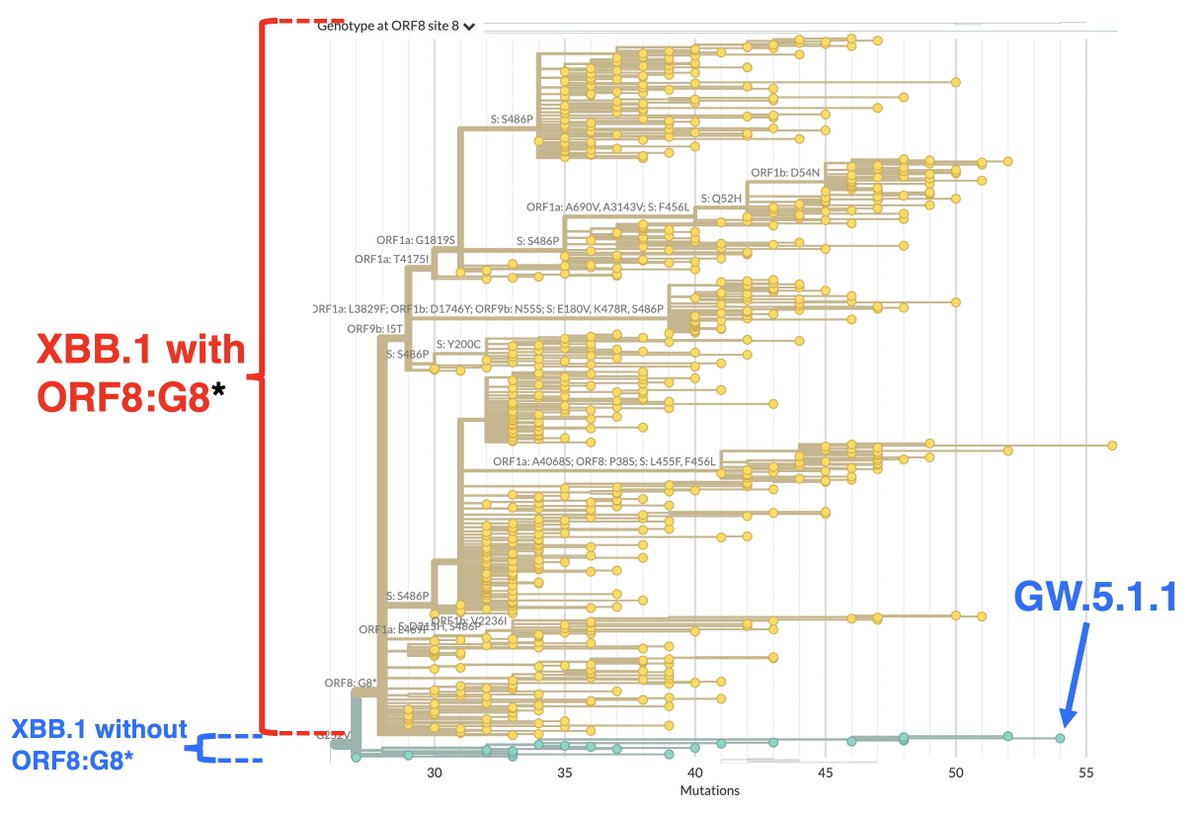
First a question: Since the ORF8 protein has been axed in the vast majority of viruses, why hasn’t the ORF8 gene been deleted from the genome? The transcription of each genome & 8/10 subgenomic mRNAs require copying the dead, 366-nuc ORF8 gene—a real waste of resources. 22/45 

One possible reason the ORF8 gene still hangs about, despite being saddled with a stop codon, is explained by pre-SARS-CoV-2 coronavirologist expert @wanderer_jasnah: Such a large deletion might scramble the secondary RNA structure. 23/45
https://twitter.com/wanderer_jasnah/status/1645518742665789446
When we talk about mutations, we're normally concerned with the amino acid (AA) change they cause, which in turn can affect the function of the protein (by making it less vulnerable to antibodies, for example). But even silent mutations (which do not cause an AA change)… 24/45 
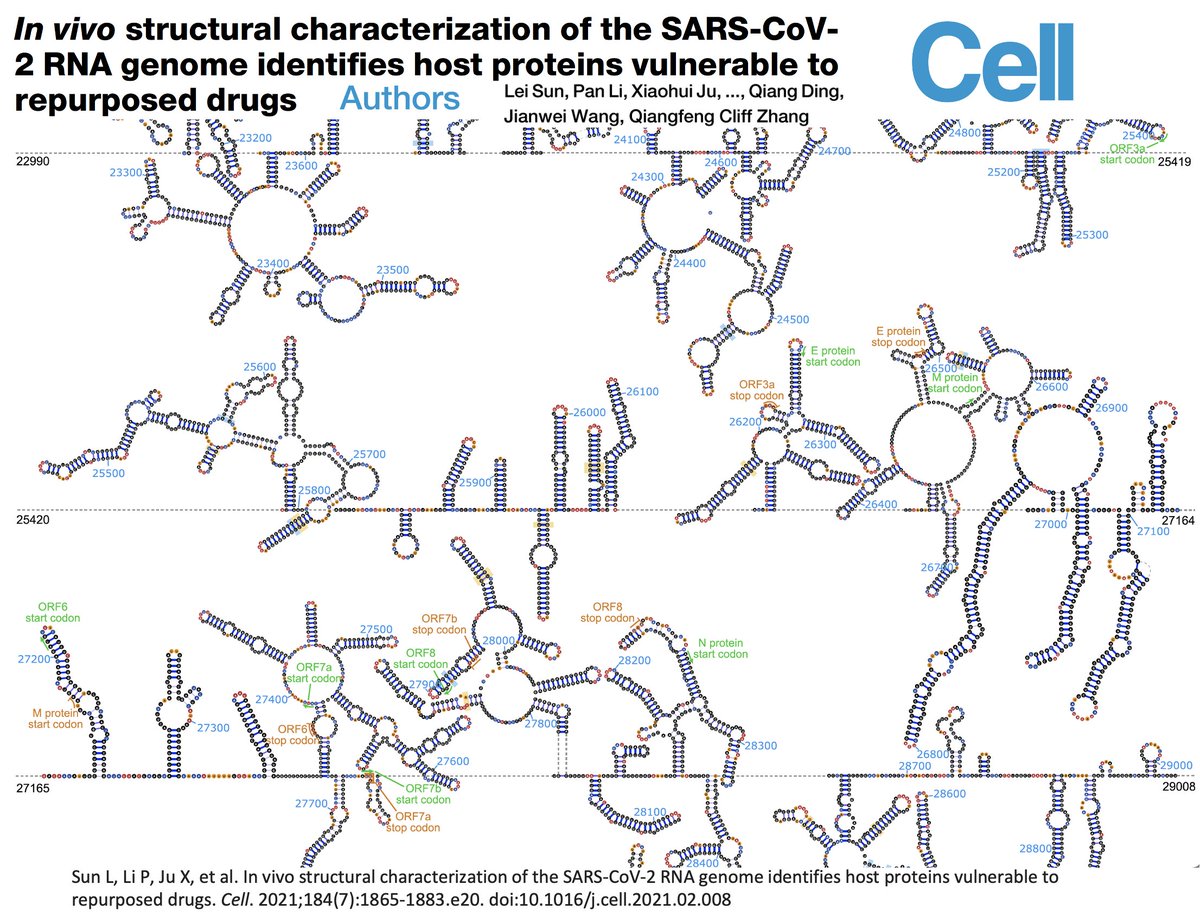
…can affect secondary RNA structure, which is the way the RNA genome twists, turns, loops, & pairs with itself. Some parts of this structure are absolutely essential, such as the stem-loops at the beginning of the genome (the 5’ UTR) shown below. 25/45 
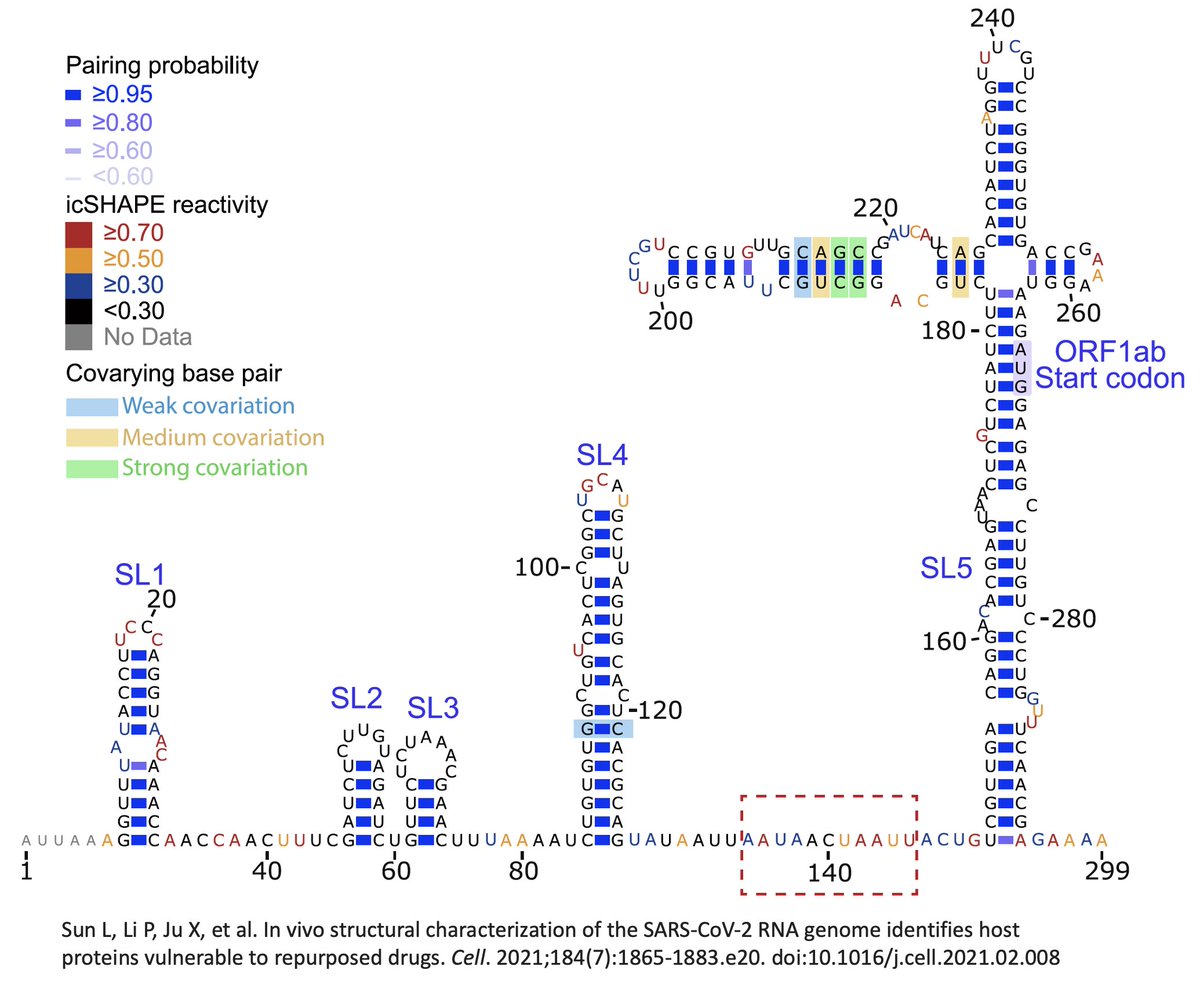
Most relevant here is the RNA structure around each subgenomic RNA TRS—7 of 8 TRS’s are located on stem-loops, & all but one face “upward,” i.e. toward the loop at the end of the stem. I don’t think the precise reason for this is known, but you can bet it’s important. 26/45 
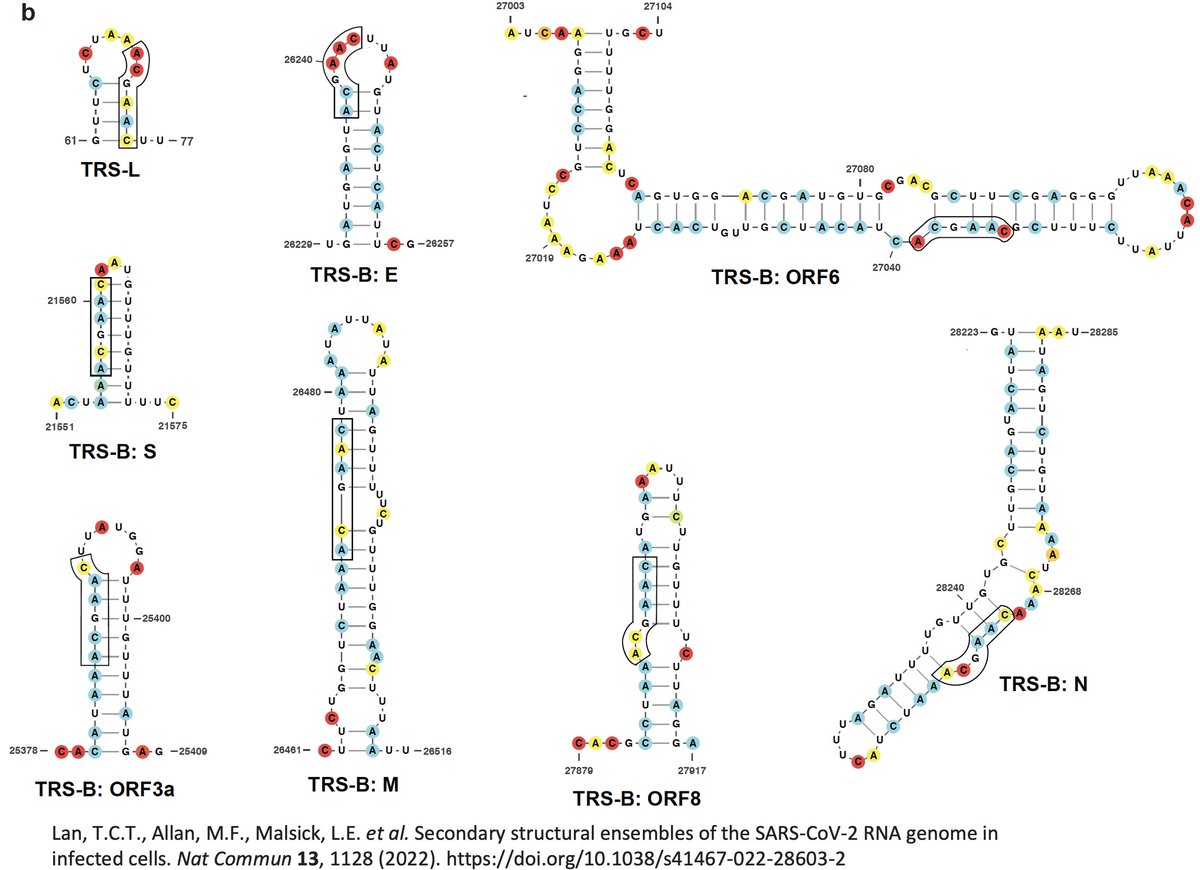
As Jasnah explains, the N/ORF9b TRS is likely the most important of all, so any deletion of ORF8 would likely need to preserve the secondary RNA structure characterizing this region. 27/45
https://twitter.com/wanderer_jasnah/status/1645518753965244433
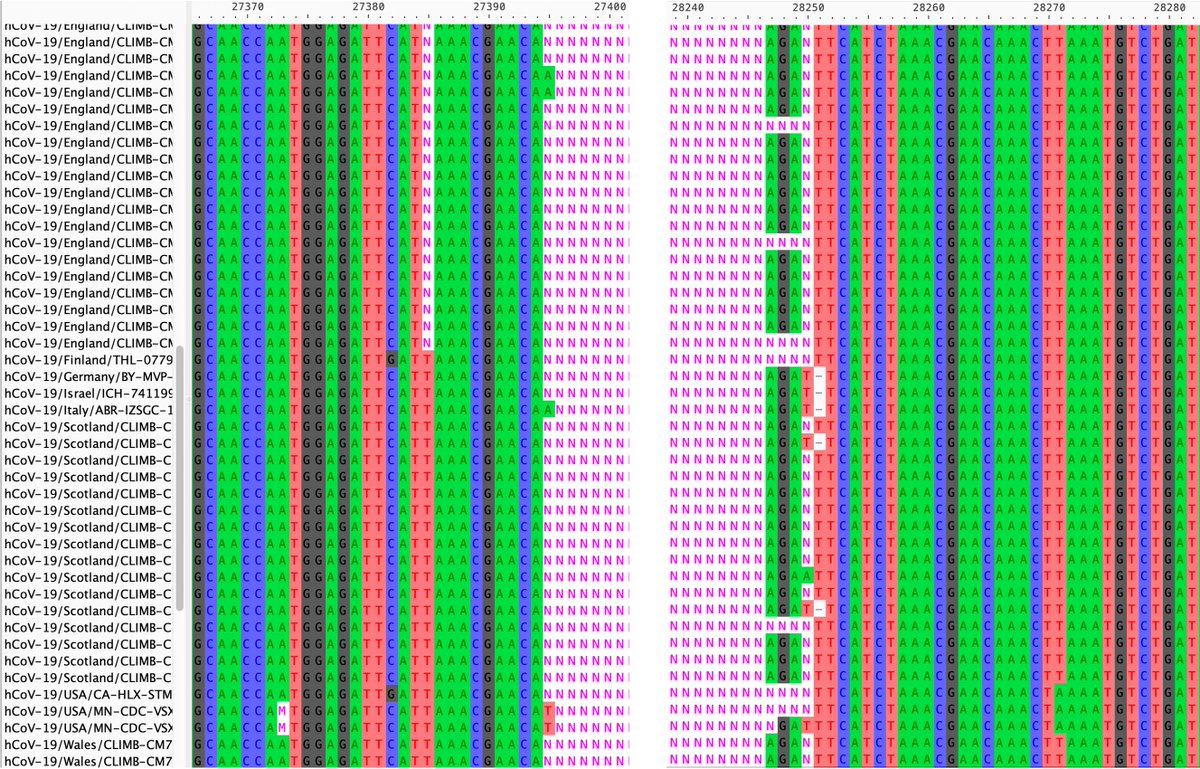
While nothing has been confirmed yet, I believe a large branch of GW.5.1.1 likely has a deletion spanning ORF7a, ORF7b, and ORF8. Nearly every genome in this branch has NNN’s (which signify “unknown”) starting just before ORF7a & ending just after ORF8. 28/45 

The deletion is so large & unexpected it confuses sequencing software, hence the NNN’s. Aligning genomes from this GW.5.1.1 branch with the invaluable Nextclade & viewing on Aliview, sequences from different countries & labs mostly match up well. 29/45 
Similarly, using the NIH’s BLAST (Basic Local Alignment Search Tool), sequences from various labs appear remarkably similar, with near-identical stretches of NNN’s. (Some sequences, mostly from labs known for less-than-perfect sequencing, differ somewhat.) 30/45



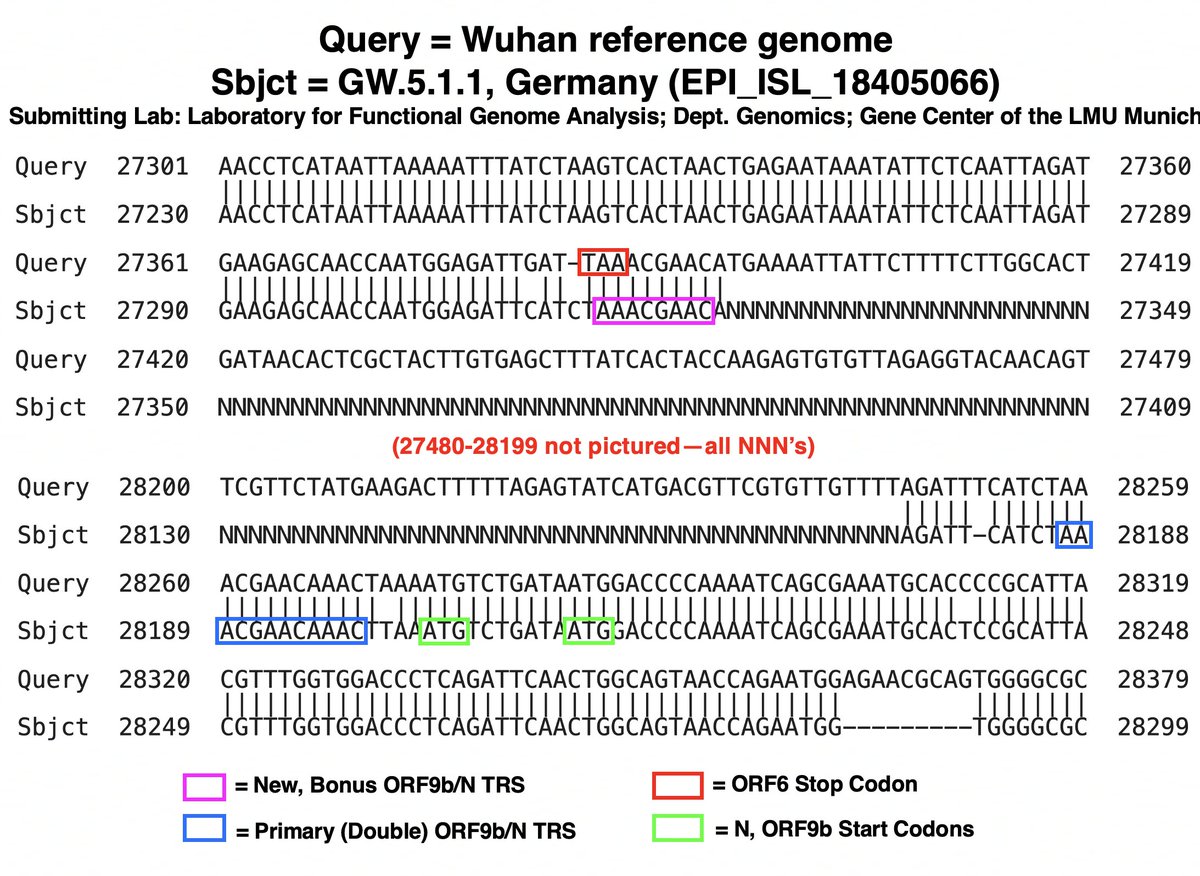
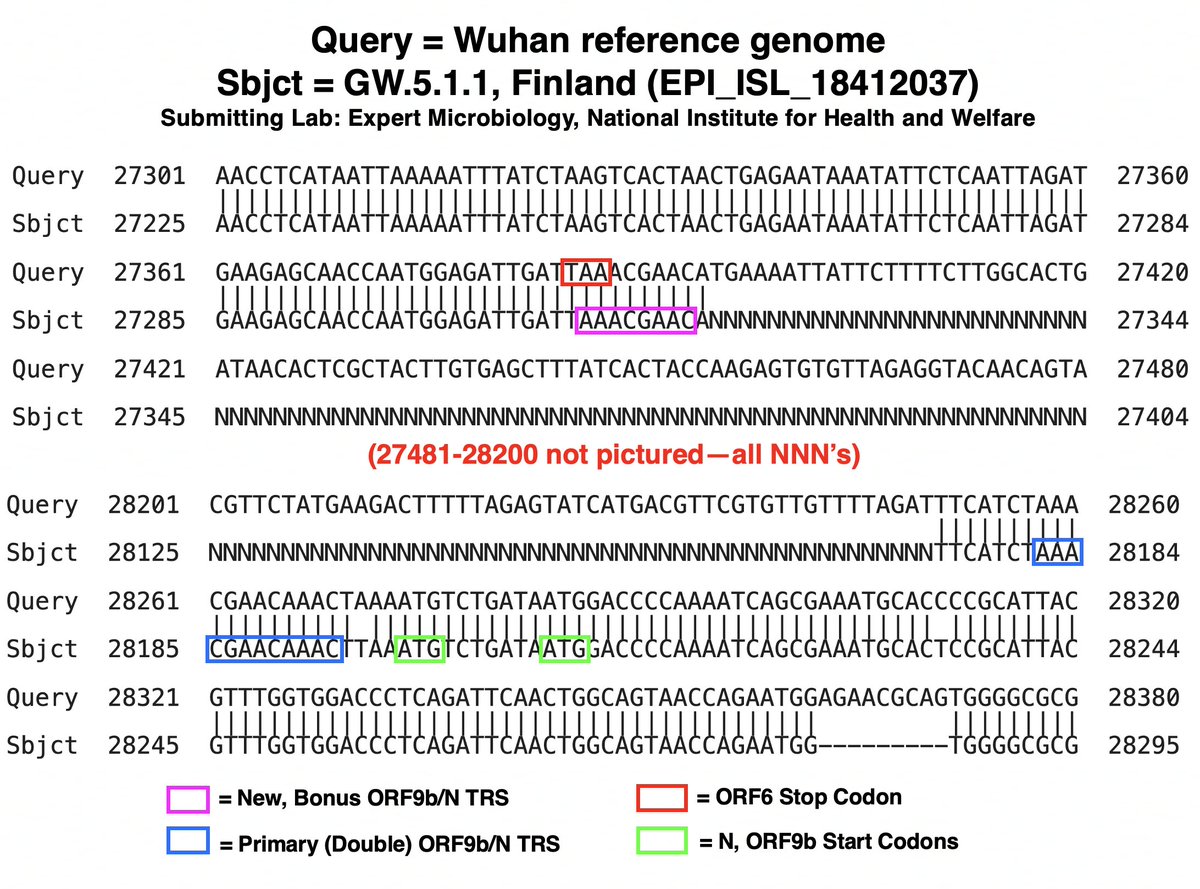
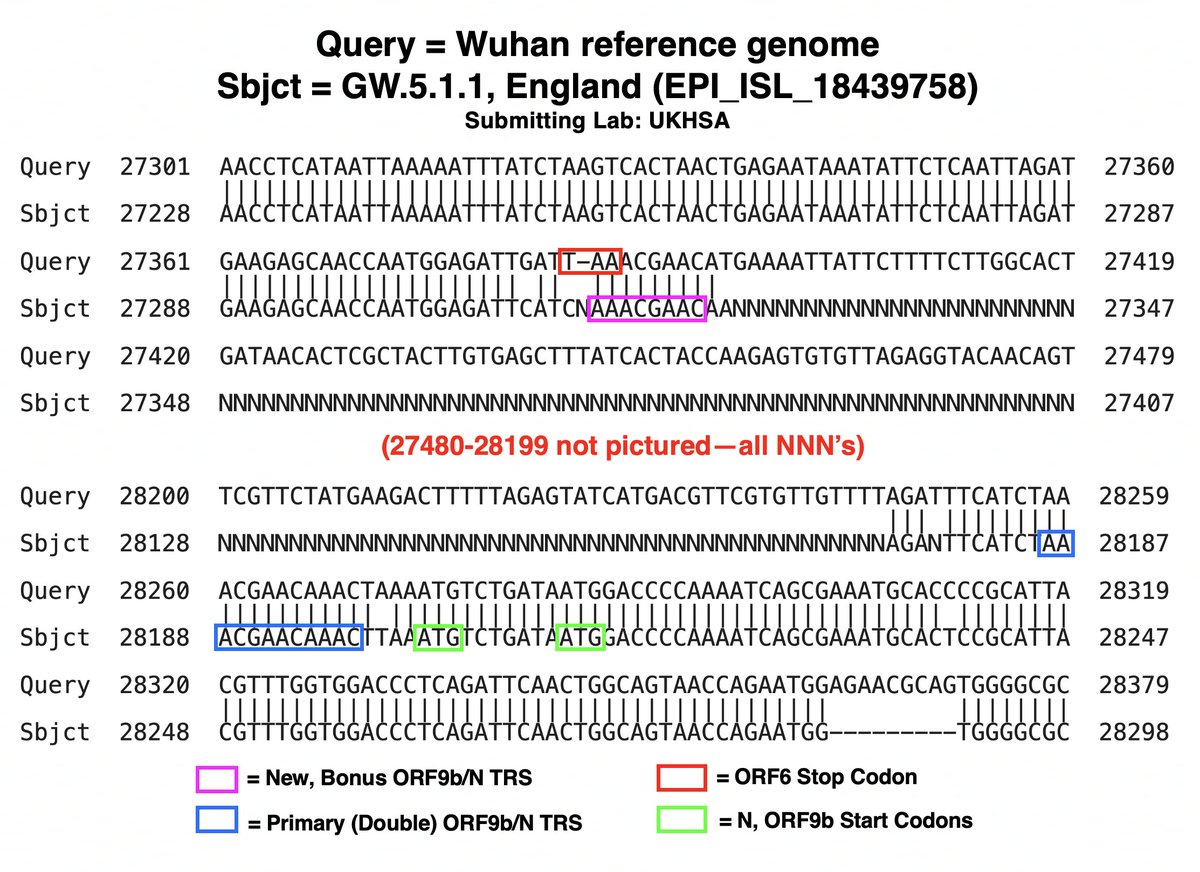
Assuming this is, in fact, a deletion—not yet a certainty—its location may not perfectly coincide with the NNN’s, as this can be affected by various sequencing technicalities (primers, etc). 31/45
But if the stretch of NNN’s roughly coincides with the posited GW.5.1.1 deletion, something remarkable occurs: the recently departed ORF7a’s TRS is hacked off & shamelessly stolen by N/ORF9b! (N & ORF9b share the same TRS.) 32/45 
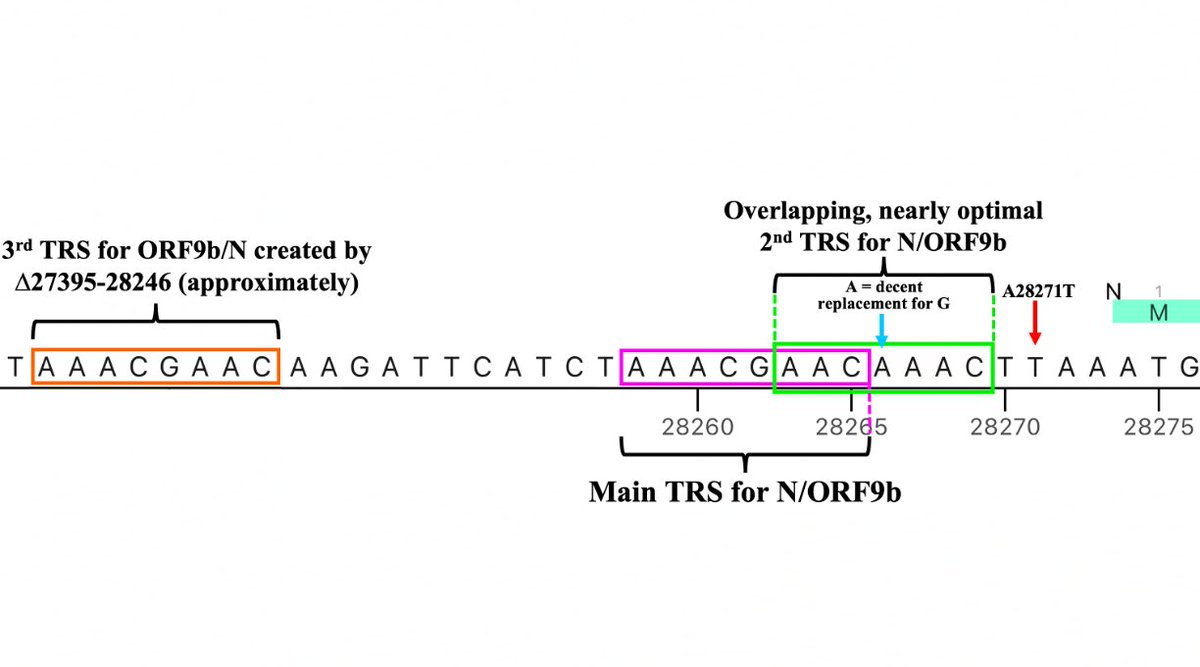
In my ORF9b 🧵, I argued that a major theme in SARS-2 evolution has been recurring mutations that increase N/ORF9b expression—especially ORF9b, whose voracious appetite for TRS is infamous. The theft of ORF7a’s TRS—if true—takes this to a new level. 33/45
https://twitter.com/LongDesertTrain/status/1673488090957004802
I actually consider this posited new TRS to be the 3rd one for N/ORF9b, the second consisting of a slightly suboptimal TRS with impressive extended homology on its 3’ end (right side)—4/5 and 8/11 are perfect matches for the 3’ end of the TRS-L. 34/45 
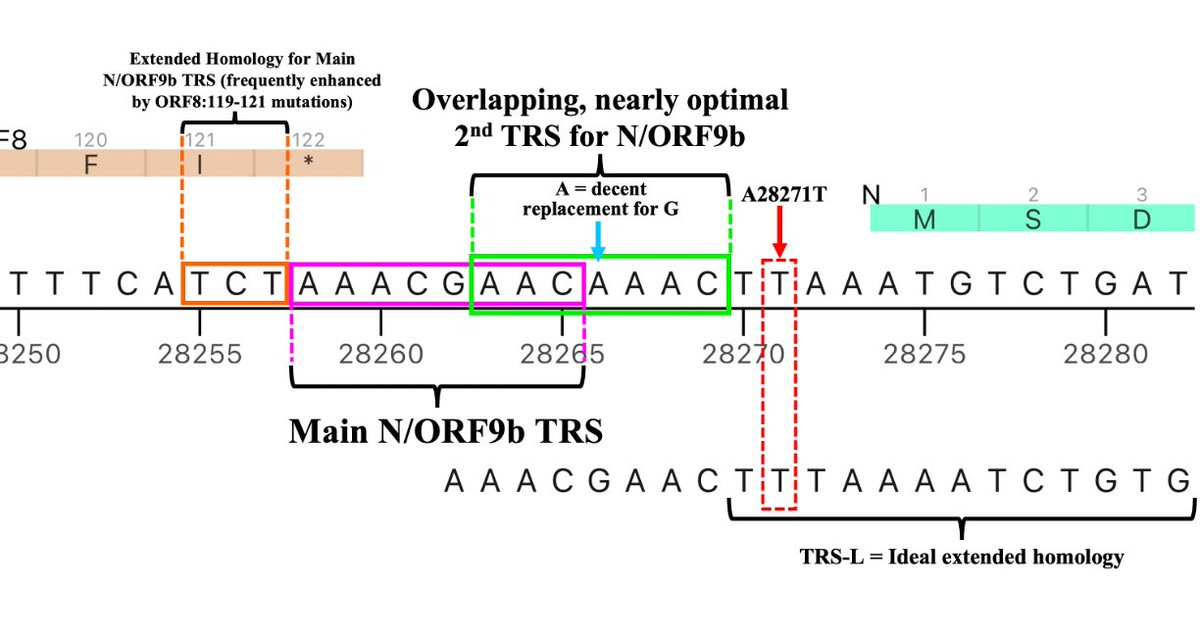
Adding something like a 3rd TRS for N/ORF9b, however, is not *entirely* unprecedented. Gamma had a 4-nucleotide insertion in the middle of the N/ORF9b double TRS that, astonishingly, added a third, overlapping TRS. 35/45 
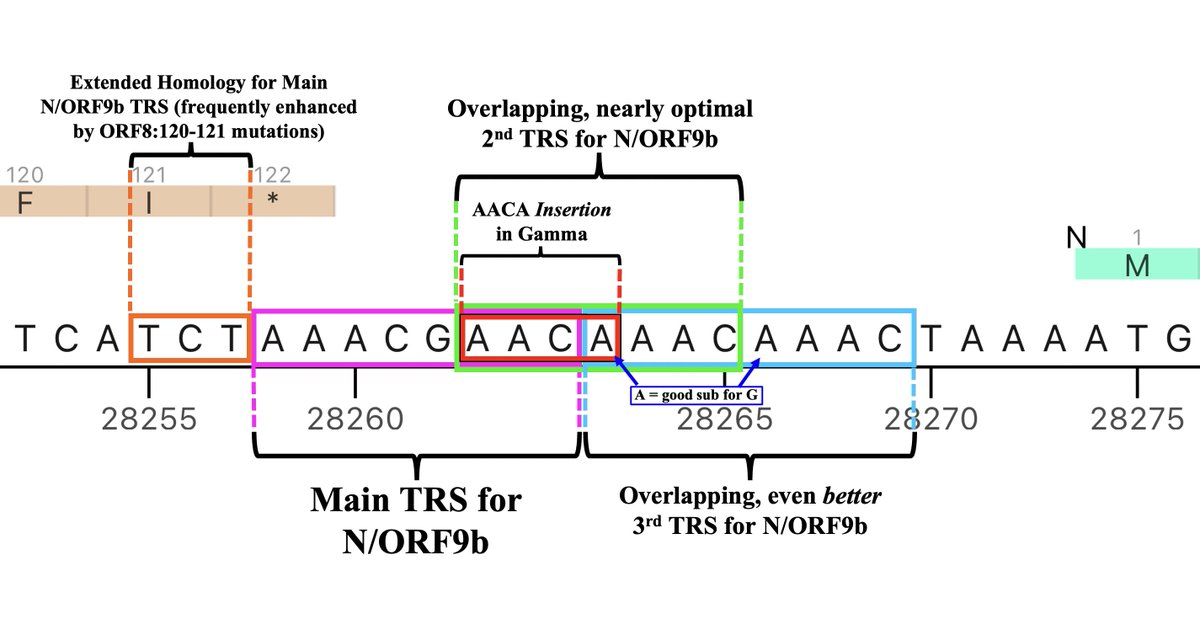
But what about the evidence, discussed above, that for lineages with the ORF6-crippling D61L, an intact ORF7a is essential? In yet another astounding twist, this GW.5.1.1 deletion has radically changed ORF6:61—either by reverting 2 nucs to form ORF6:L61H… 36/45 
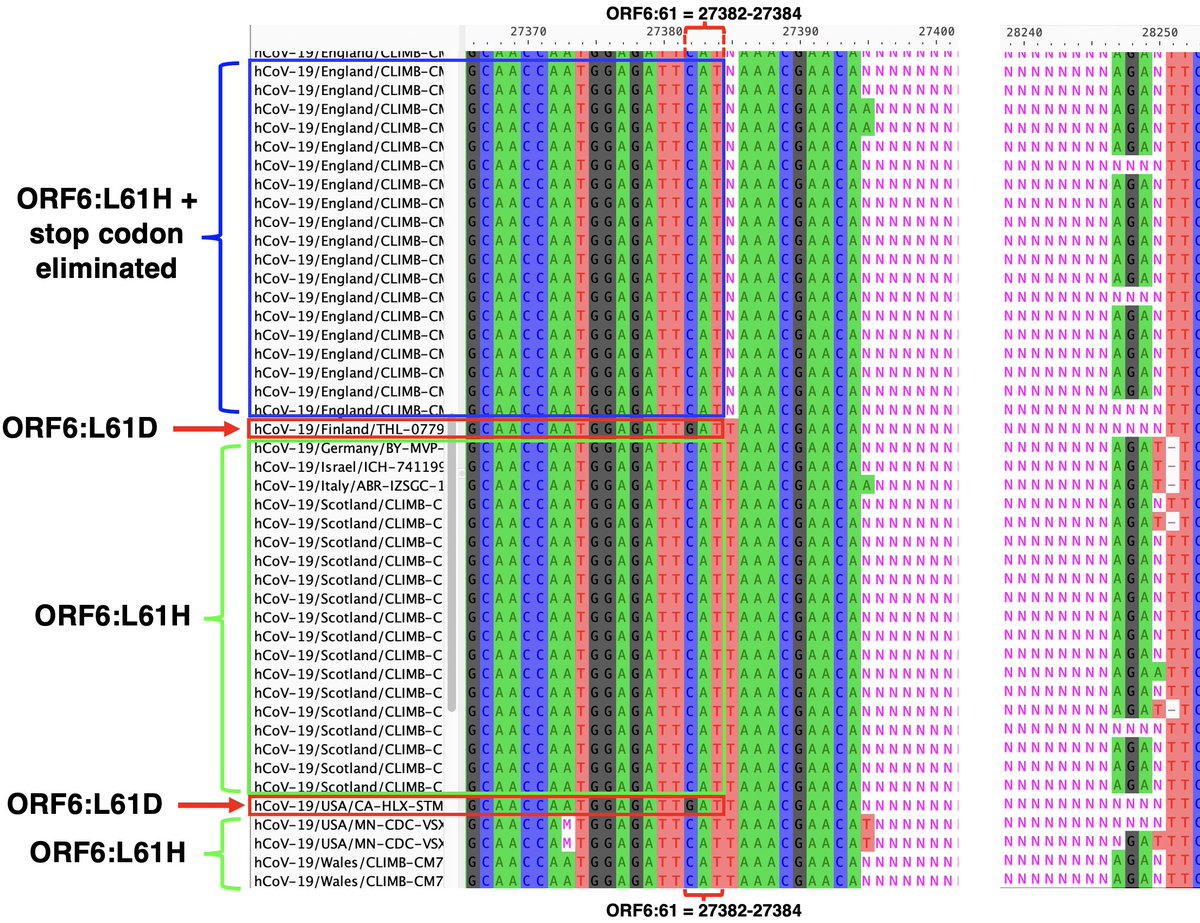
…reverting entirely to ORF6:L61D, or maybe by trashing the stop codon & extending ORF6 by another. It’s not 100% clear what’s happened at ORF6:61 because the edges of the posited deletion are messy, but most sequences imply some major change has occurred. 37/45 
GW.5.1.1 is not the only new lineage that appears to possess large deletions across ORF7ab-ORF8. At least two others, probably more, have similar, but somewhat shorter, deletions. One branch of EG.5.1, e.g., features NNN’s across ORF8 & half of ORF7b. 38/45 
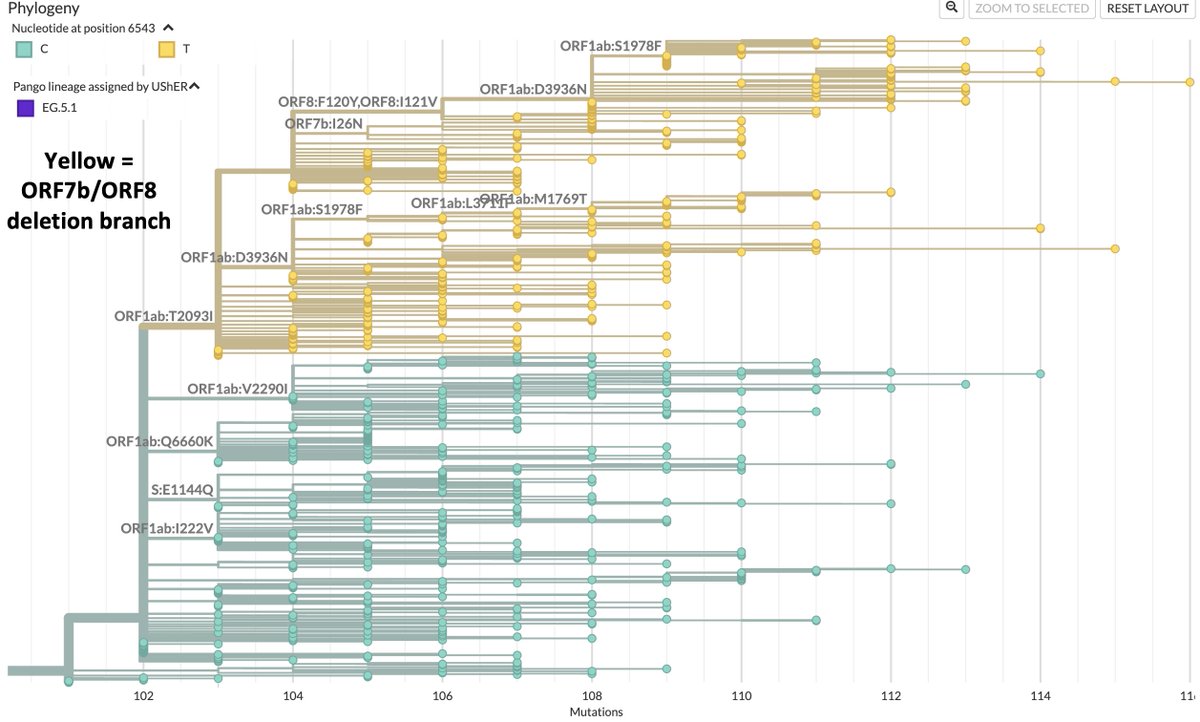
Dozens of sequences in this branch from Japan actually register a deletion where others only have NNN’s, but the resulting mutant is too heinous for Nextclade to contemplate. 39/45 
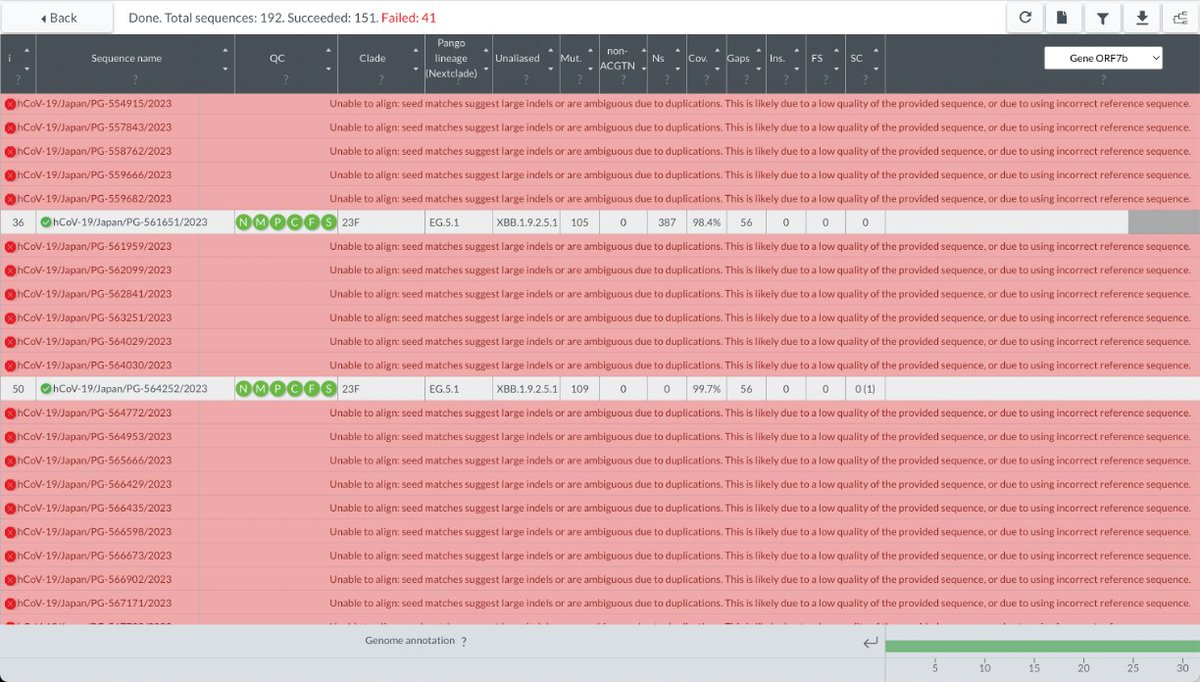
BLAST, however, looks the hideous mutant straight in the eye, & seems to give a truthful report. 40/45 
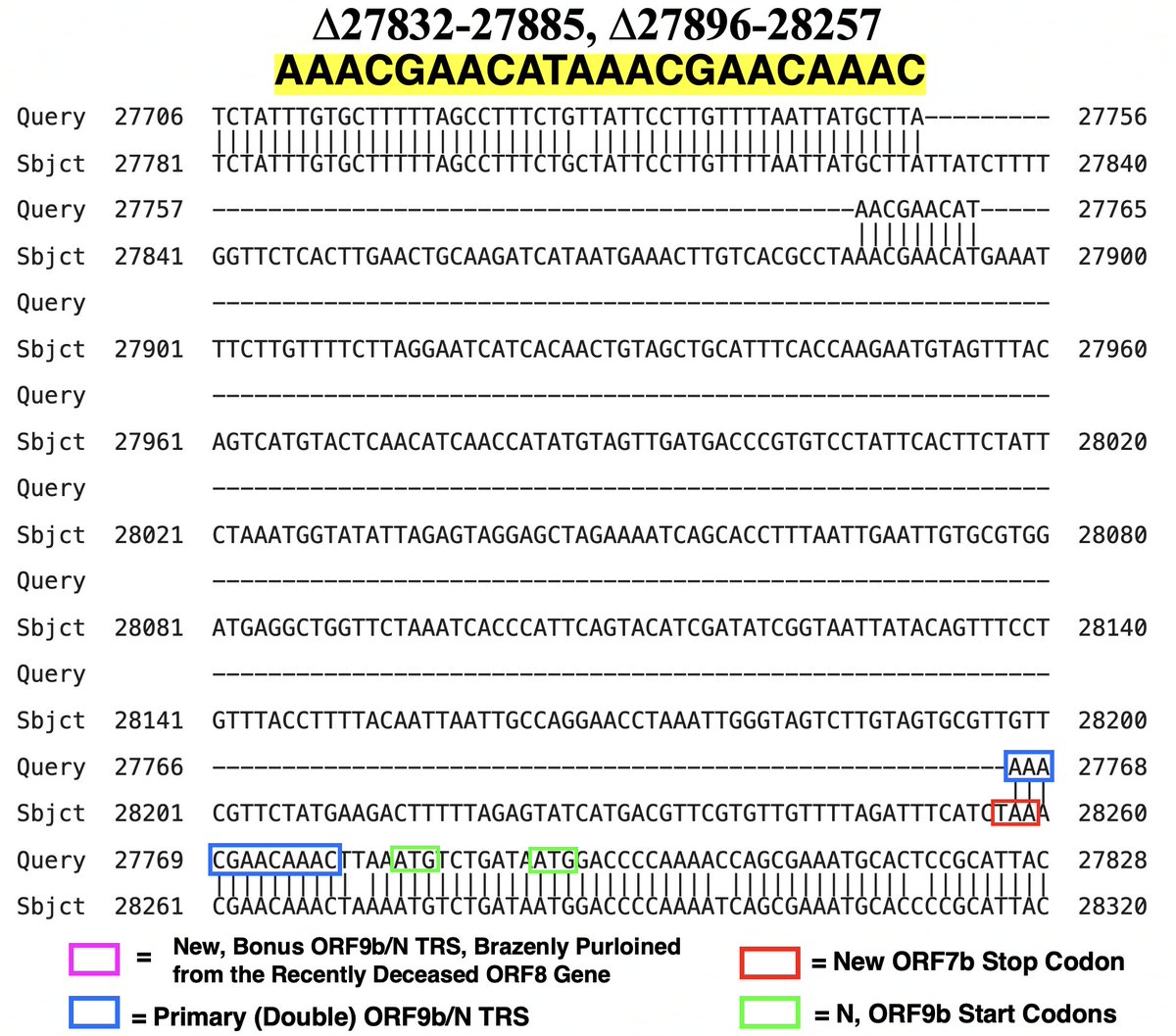
ORF7a is spared, while half of ORF7b is lost. The kicker? This deletion *also* seems to add a 3rd TRS for ORF9b/N. This time N/ORF9b, like a graverobber swiping the boots from a corpse, pilfers the TRS of the newly deceased ORF8 gene. 41/45 
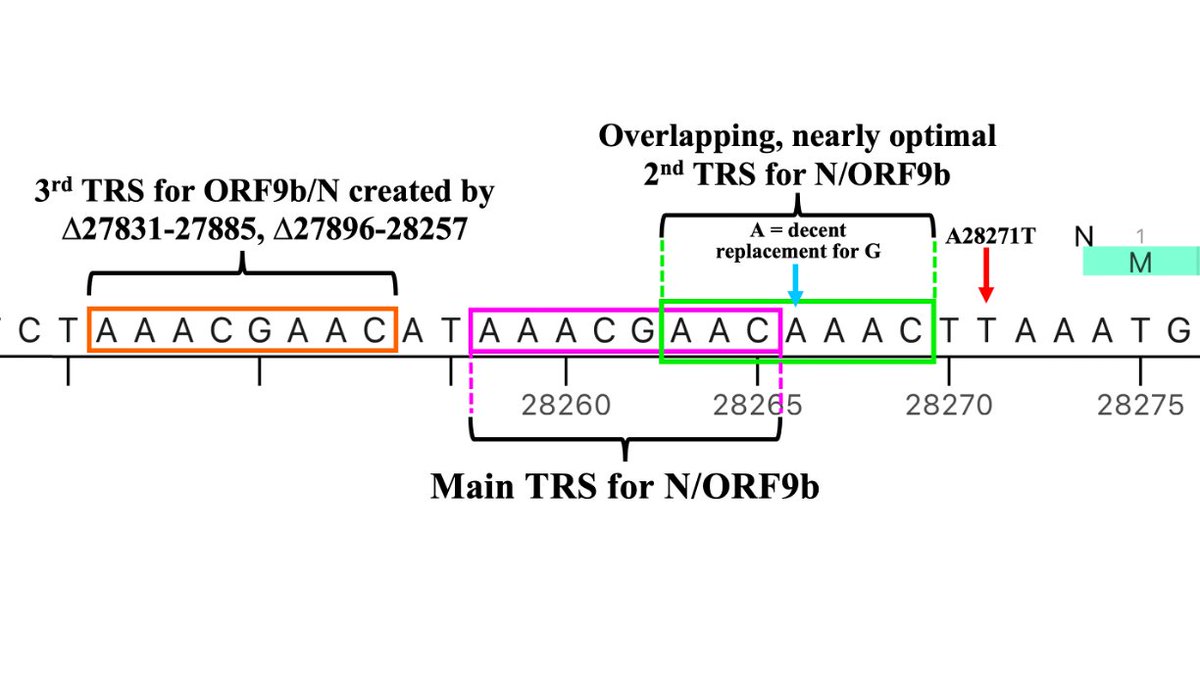
I want to emphasize that while I strongly suspect something like what I’ve described above is going on, nothing has been verified or is certain. It’s possible something entirely different is happening. We await word from the oracular readers of raw sequences. 42/45
But assuming it is real, what does it all mean? Possibly nothing. After all, this is only a very small fraction of sequences, & like various other peculiarities—such as the various double-FCS lineages with S:P812R—this may be a lot of sound & fury, signifying nothing. 43/45
Seeing similar deletions in multiple branches suggests changing selection pressures & reinforces the impressive ability of SARS-CoV-2 to refashion itself & the real possibility of future abrupt genomic changes. But the precise real-world consequences? Impossible to predict. 44/45
Before ending, let’s acknowledge the impressive foresight of @wanderer_jasnah, who predicted ORF8 loss & eventual deletion (see above: tweets 23, 27) & @trvrb, whose recent ORF8 knockout paper suggested ORF7a, ORF7b, & ORF8 are prime candidates for deletion. 45/end 
Thank you:
• @theosanderson for making GenSplore, an essential tool used in many of the diagrams above (whose only flaw is the lack of ORF7b labeling)
• @ChaoranChen_ for creating & continually improving the amazing Cov-Spectrum
• @theosanderson for making GenSplore, an essential tool used in many of the diagrams above (whose only flaw is the lack of ORF7b labeling)
• @ChaoranChen_ for creating & continually improving the amazing Cov-Spectrum
Thank you:
• Lab workers, who do the groundwork without which any kind of analysis would be impossible—esp. sequencers in Japan in this case.
• @DarrenM98230782, @PeacockFlu, @siamosolocani, @Asinickle, @nzm8qs, @JosetteSchoenma, @gianlucac1, @shay_fleishon, @wolfeagle1989
• Lab workers, who do the groundwork without which any kind of analysis would be impossible—esp. sequencers in Japan in this case.
• @DarrenM98230782, @PeacockFlu, @siamosolocani, @Asinickle, @nzm8qs, @JosetteSchoenma, @gianlucac1, @shay_fleishon, @wolfeagle1989
Thank you:
• Lab workers, who do the groundwork without which any kind of analysis would be impossible—esp. sequencers in Japan in this case.
• @DarrenM98230782, @PeacockFlu, @siamosolocani, @Asinickle, @nzm8qs, @JosetteSchoenma, @gianlucac1, @shay_fleishon, @wolfeagle1989
• Lab workers, who do the groundwork without which any kind of analysis would be impossible—esp. sequencers in Japan in this case.
• @DarrenM98230782, @PeacockFlu, @siamosolocani, @Asinickle, @nzm8qs, @JosetteSchoenma, @gianlucac1, @shay_fleishon, @wolfeagle1989
• • •
Missing some Tweet in this thread? You can try to
force a refresh