BREAKING🔔 A new study from G2P-Japan🇯🇵 is out at bioRxiv @biorxivpreprint. We elucidated the virological characteristics of SARS-CoV-2 BA.2.86 (aka #Pirola).
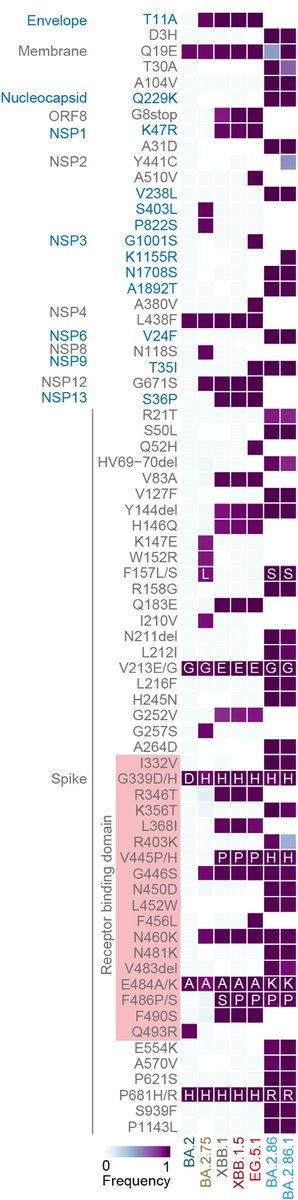
*BA.2.86 is has >30 mutations in the genome.
Please RT🔥 1/
biorxiv.org/content/10.110…


*BA.2.86 is has >30 mutations in the genome.
Please RT🔥 1/
biorxiv.org/content/10.110…
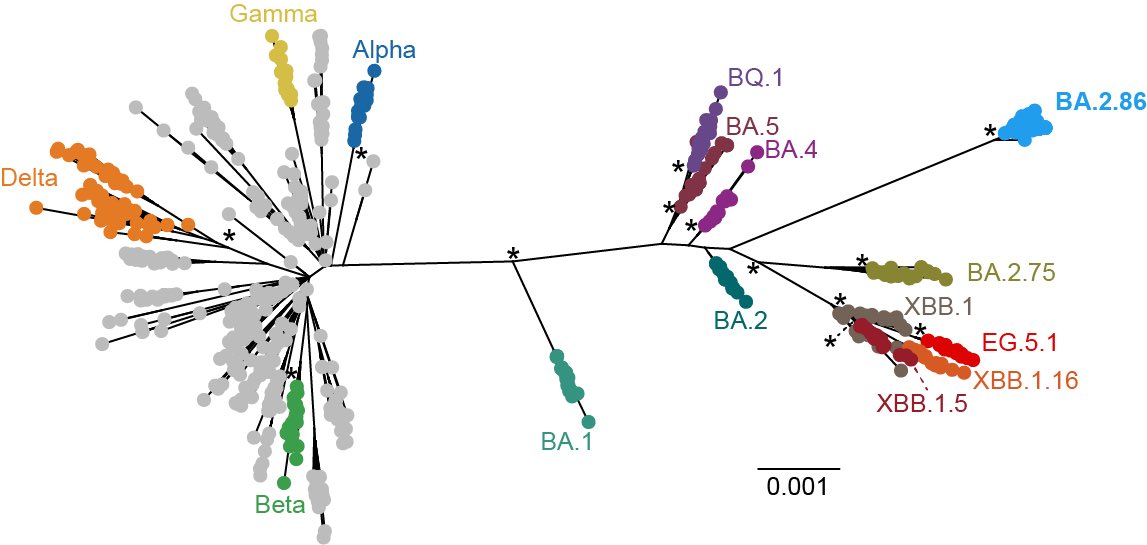
Epidemic dynamics modeling by @jampei2 and @JarelElgin showed that the relative effective reproduction number (Re) of BA.2.86 is 1.07-fold higher than that of the currently predominant EG.5.1, suggesting that BA.2.86 has the potential to become the next predominant one. 2/ 

Growth assay showed that the replication capacity of BA.2.86 is similar to or lower than that of BA.2, its parental strain. BA.2.86 is less replicative than EG.5.1 at the cell culture level. 3/ 

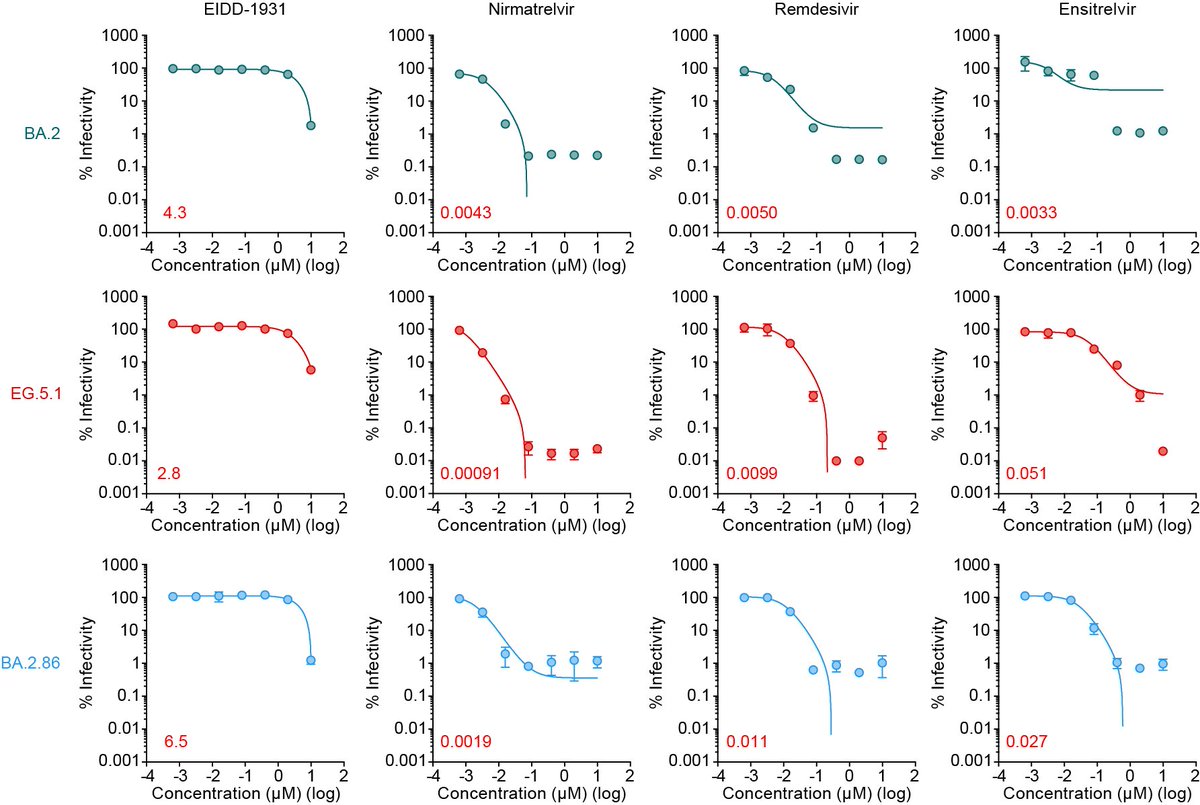
Four antivirals, molnupiravir (EIDD-1931), nirmatrelvir, remdesivir and ensitrelvir were all effective against BA.2.86. 4/ 
A cell-based fusion assay showed that the fusogenicity of BA.2.86 spike is a bit (1.5-fold) greater than that of the parental BA.2 spike. 5/ 

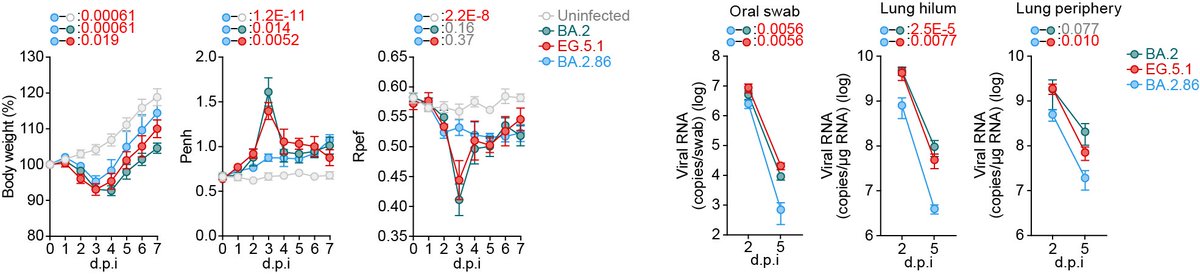
Infection experiments in hamsters showed that both the intrinsic pathogenicity and replication kinetics of BA.2.86 are significantly lower than those of the parental BA.2. 6/ 
Overall, BA.2.86 does not appear to pose an increased risk to human society. However, it is possible that its virological phenotype may change or even worsen in the future by acquiring additional mutations. Continued monitoring and surveillance are important now and then. 7/7
• • •
Missing some Tweet in this thread? You can try to
force a refresh

 Read on Twitter
Read on Twitter










