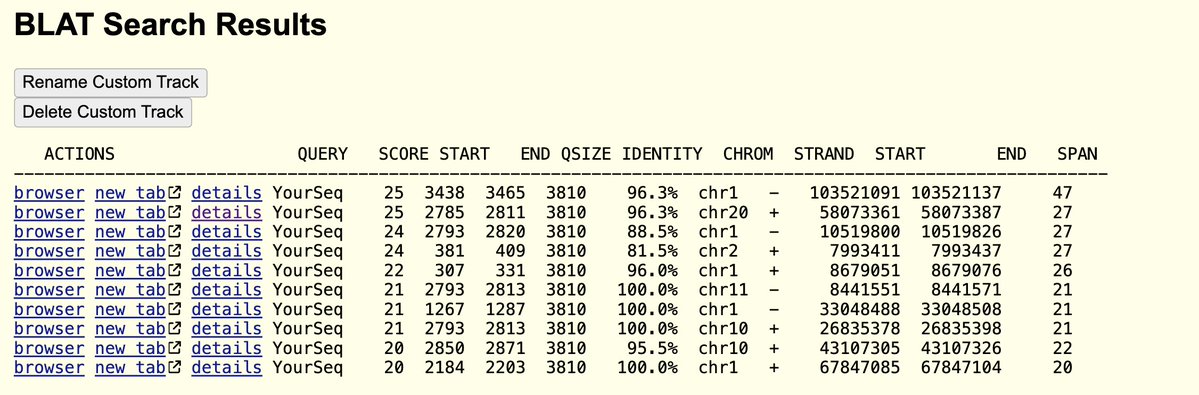
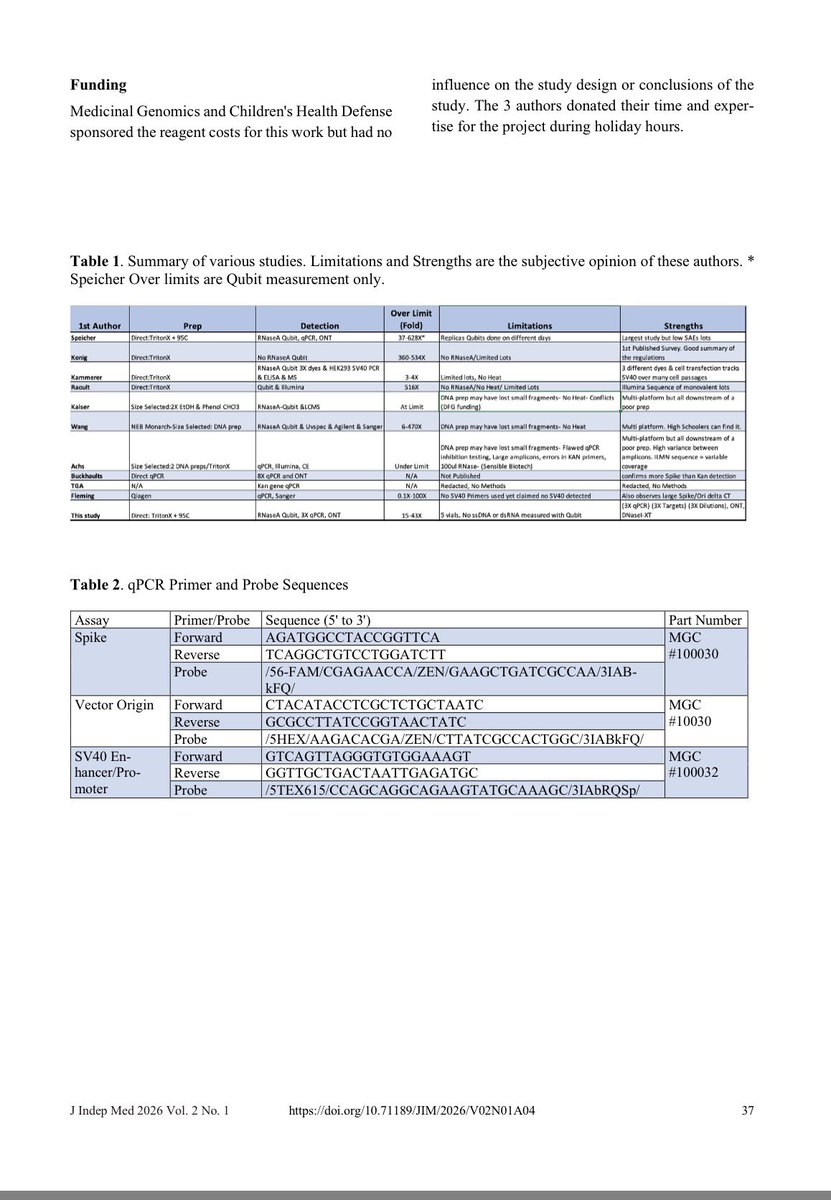
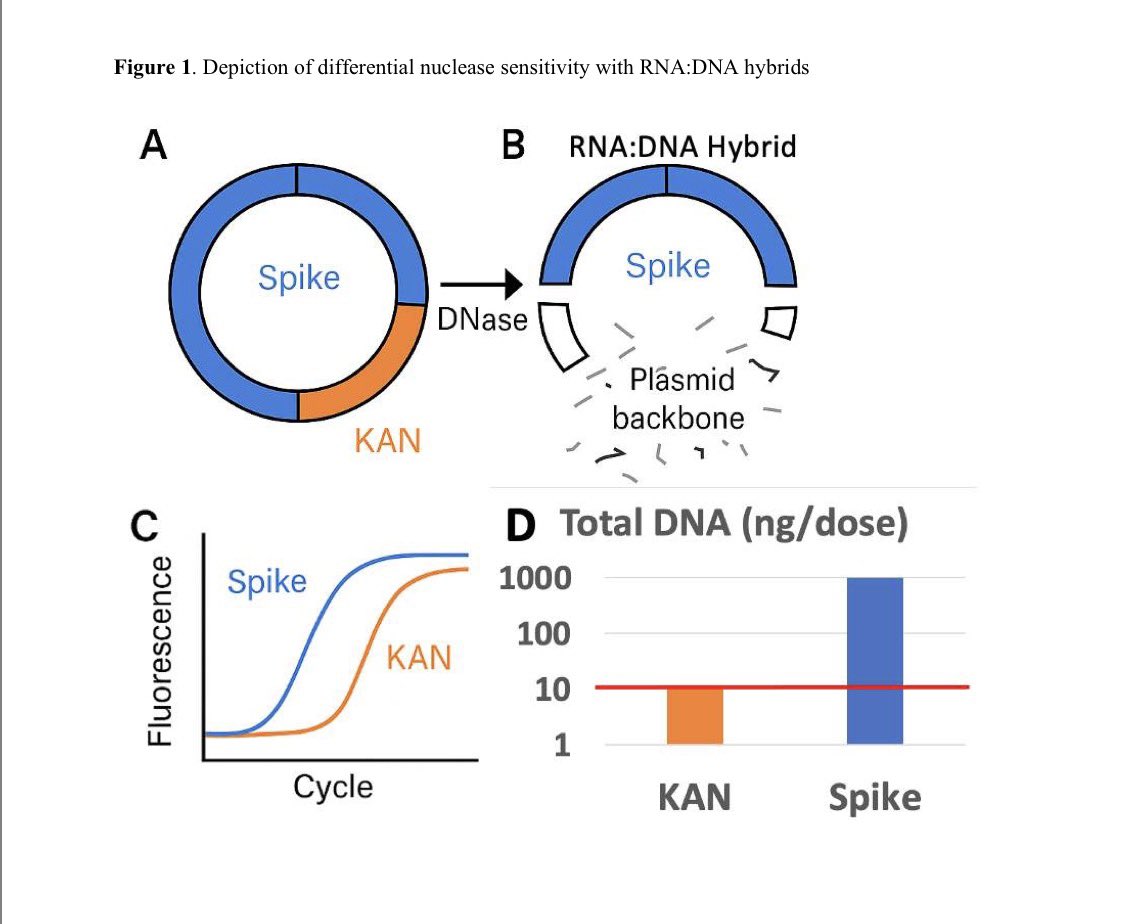
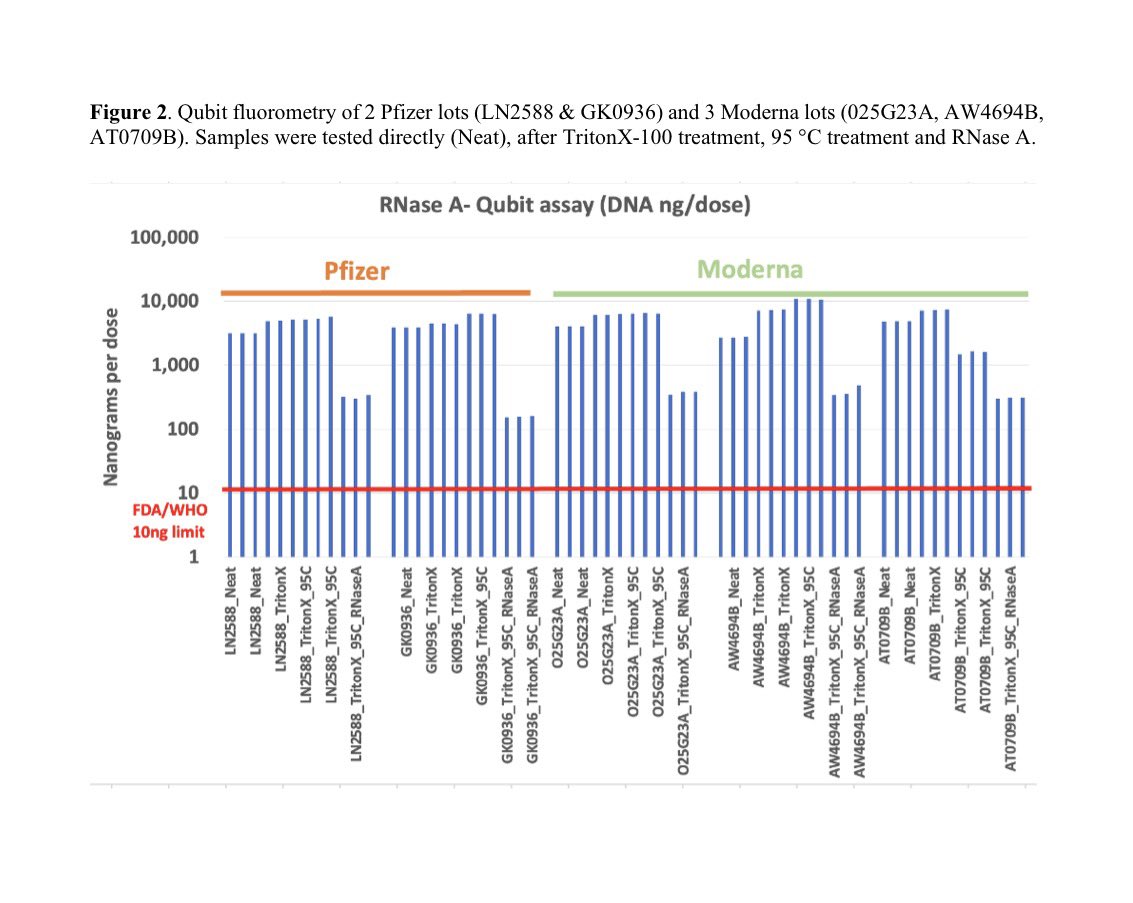
A reminder that this DNA has SV40 origins of replication.
This sequence binds to p53 and heads to the nucleus.
anandamide.substack.com/p/shrodingers-…
This sequence binds to p53 and heads to the nucleus.
anandamide.substack.com/p/shrodingers-…
• • •
Missing some Tweet in this thread? You can try to
force a refresh