BREAKING🔔 A new study from G2P-Japan🇯🇵 is out at bioRxiv @biorxivpreprint. We elucidated the virological characteristics of SARS-CoV-2 JN.1 - a progeny of BA.2.86 (#Pirola). Please RT🔥 1/
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
Cf. This is JN.1 (aka BA.2.86.1.1; in spike, JN.1 = BA.2.86 + S:L455S. Importantly, we remember that the L455 substitution, S:L455F, was observed in the XBB lineage (e.g., HK.3 and these are known as "FLip" subvariants). 2/ 

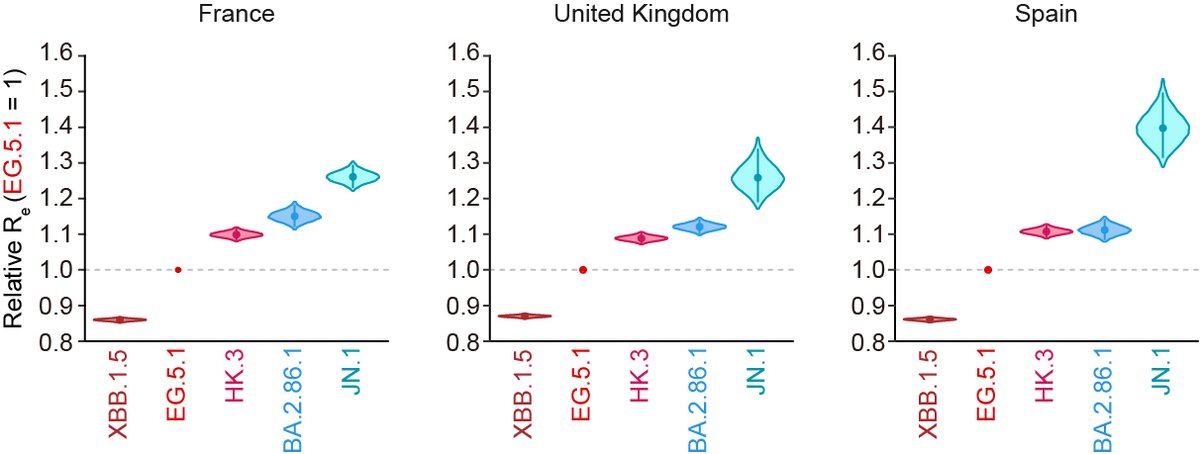
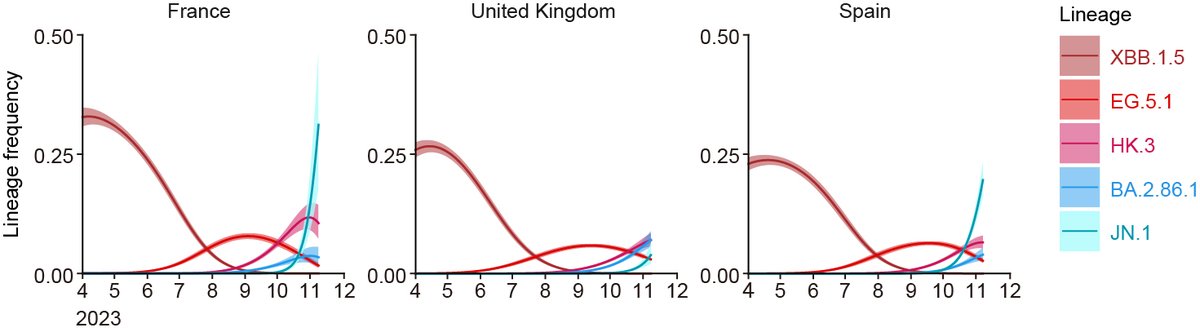
Transmissibility/fitness (Re):
Molecular phylogenetic-epidemic dynamics modeling led by Jumpei @jampei2 and Kaho showed that the Re of JN.1 is apparently greater than that of BA.2.86 and others, suggesting that S:L455S can contribute to the increased fitness of JN.1. 3/


Molecular phylogenetic-epidemic dynamics modeling led by Jumpei @jampei2 and Kaho showed that the Re of JN.1 is apparently greater than that of BA.2.86 and others, suggesting that S:L455S can contribute to the increased fitness of JN.1. 3/
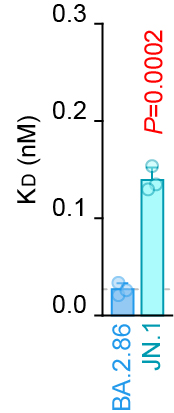
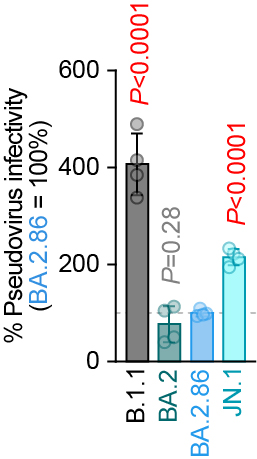
ACE2 binding & pseudovirus infectivity:
ACE2 binding: BA.2.86 < JN.1
Pseudovirus infectivity: BA.2.86 > JN.1
Effect of S:L455S is different between monomeric RBD and trimerized whole S protein🤔? 4/


ACE2 binding: BA.2.86 < JN.1
Pseudovirus infectivity: BA.2.86 > JN.1
Effect of S:L455S is different between monomeric RBD and trimerized whole S protein🤔? 4/
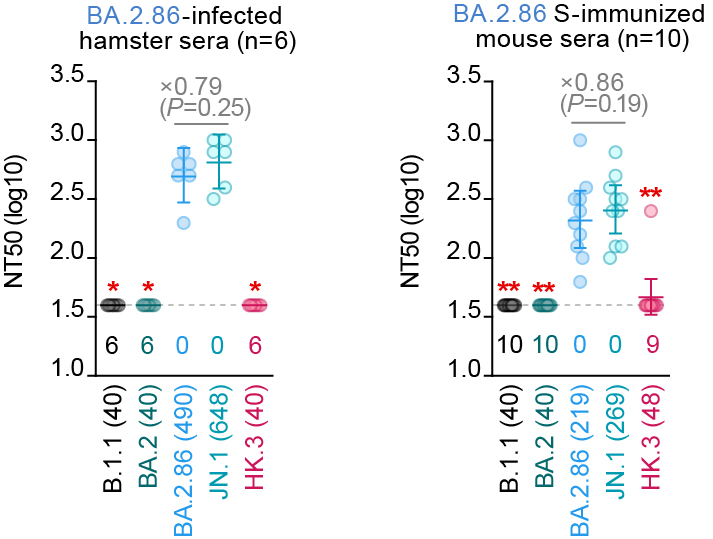
Immune resistance (rodent sera):
In cases of BA.2.86-infected hamster sera and BA.2.86 S-immunizesd mouse sera, the 50% neutralization titer (NT50) between BA.2.86 and JN.1 was comparable, suggesting that S:L455S does not affect the antigenicity of BA.2.86. 5/
In cases of BA.2.86-infected hamster sera and BA.2.86 S-immunizesd mouse sera, the 50% neutralization titer (NT50) between BA.2.86 and JN.1 was comparable, suggesting that S:L455S does not affect the antigenicity of BA.2.86. 5/
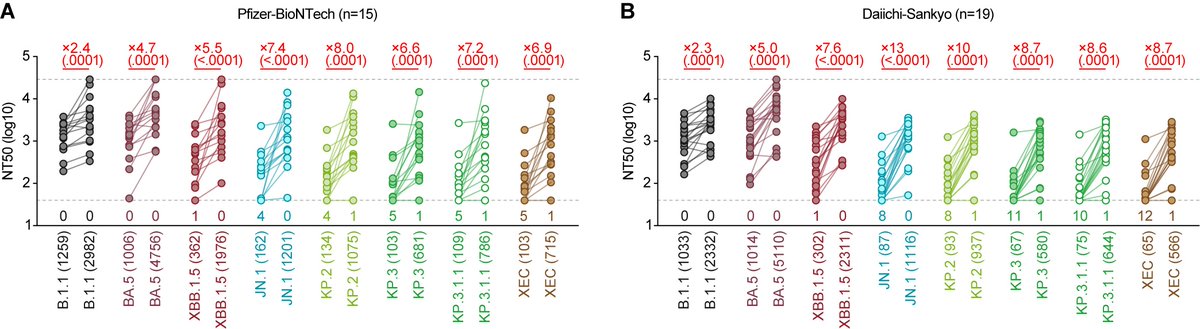
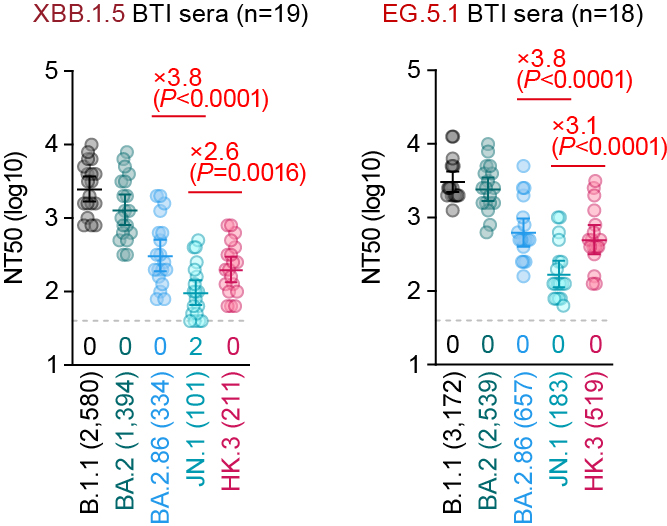
Immune resistance (human sera):
On the other hand, the NT50 of breakthrough infection (BTI) sera with XBB.1.5 and EG.5.1 against JN.1 was significantly lower than that of HK.3 (2.6- to 3.1-fold) and BA.2.86 (3.8-fold)🔥 6/
On the other hand, the NT50 of breakthrough infection (BTI) sera with XBB.1.5 and EG.5.1 against JN.1 was significantly lower than that of HK.3 (2.6- to 3.1-fold) and BA.2.86 (3.8-fold)🔥 6/
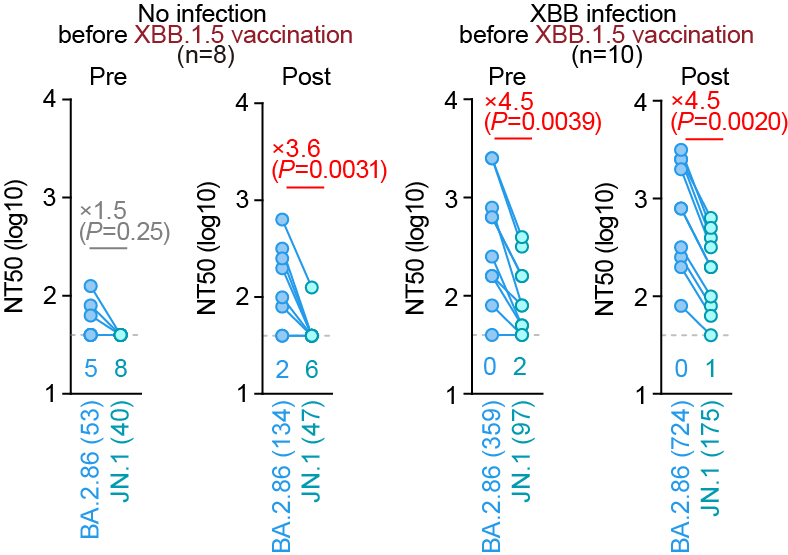
Furthermore, JN.1 shows a robust resistance to monovalent XBB.1.5 vaccine sera🔥 These results suggest that JN.1 is one of the most immune-evading variants to date. S:L455S can contribute to increased immune evasion, which partly explains the increased Re of JN.1. 7/7 
• • •
Missing some Tweet in this thread? You can try to
force a refresh