Our paper on JN.1 is now online @TheLancetInfDis!
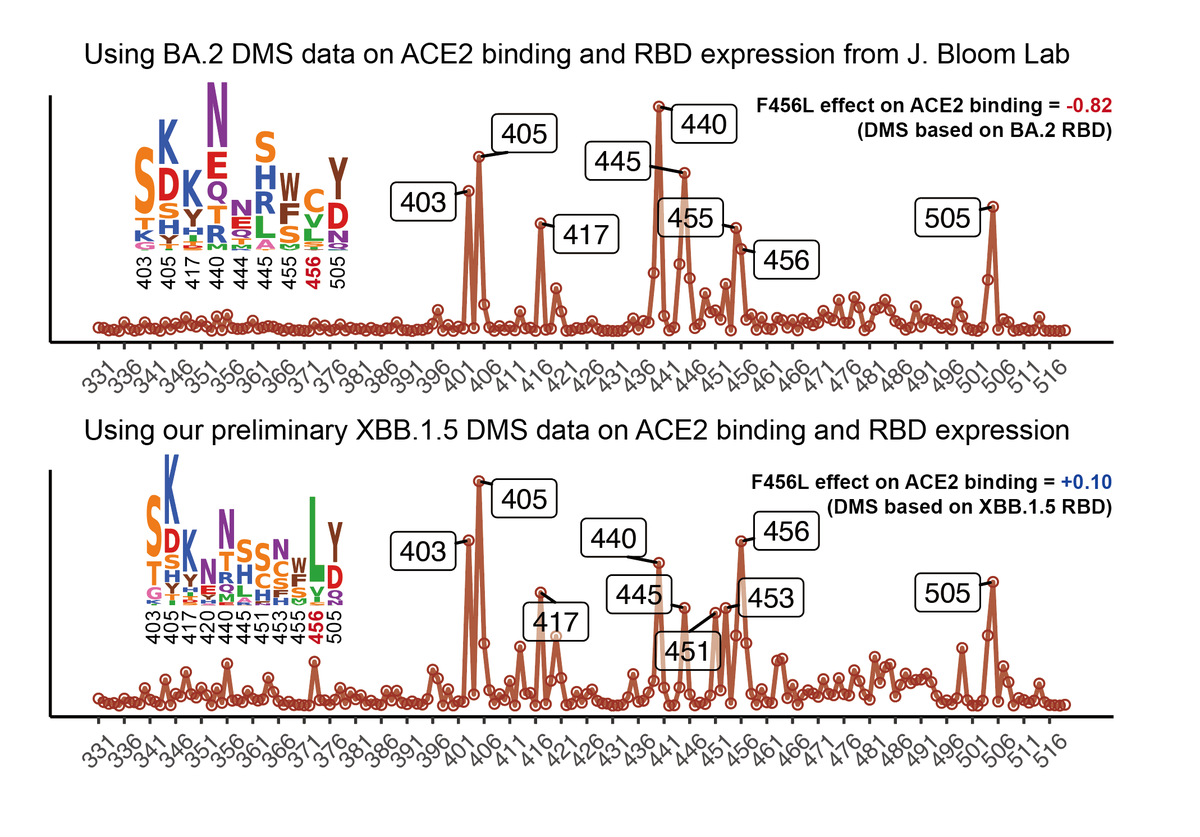
The manuscript explains how a single RBD mutation L455S could turn BA.2.86 into a heavy immune evasive variant JN.1.
Notably, JN.1 is now approaching worldwide dominance (42% two weeks ago).
thelancet.com/journals/lanin…
The manuscript explains how a single RBD mutation L455S could turn BA.2.86 into a heavy immune evasive variant JN.1.
Notably, JN.1 is now approaching worldwide dominance (42% two weeks ago).
thelancet.com/journals/lanin…
Two months ago, we warned about JN.1 due to its extreme immune evasion. The reason why we paid attention to JN.1 so early is that we know BA.2.86 is very weak to Class 1 antibodies and L455S is one of the strongest Class 1 antibody escaping mutations. 2/6
https://twitter.com/yunlong_cao/status/1714694253530804588
Many labs have shown that BA.2.86 is well-neutralized. However, the absolute neutralizing titers cannot tell the full story. Since the majority of BA.2.86-neutralizing Abs are from a single epitope, huge changes in titers could happen when BA.2.86 acquires critical mutations. 3/6
So in the future, we should not only focus on the serum neutralization titers of a new variant but also care about the characteristics of the effective neutralizing antibodies, especially when their epitope distributions are dominated by one class, similar to BA.2.86. 4/6
Also, the case of JN.1 highlights the importance of closely monitoring strains with high ACE2 binding affinity and distinct antigenicity, like BA.2.75 and BA.2.86, despite their temporarily unremarkable immune evasion capabilities. 5/6
Such strains could survive and transmit at low levels, since their large antigenic distance to dominant strains allows them to target distinct populations and accumulate immune-evasive mutations rapidly, often at the cost of receptor binding affinity. 6/6
• • •
Missing some Tweet in this thread? You can try to
force a refresh

 Read on Twitter
Read on Twitter