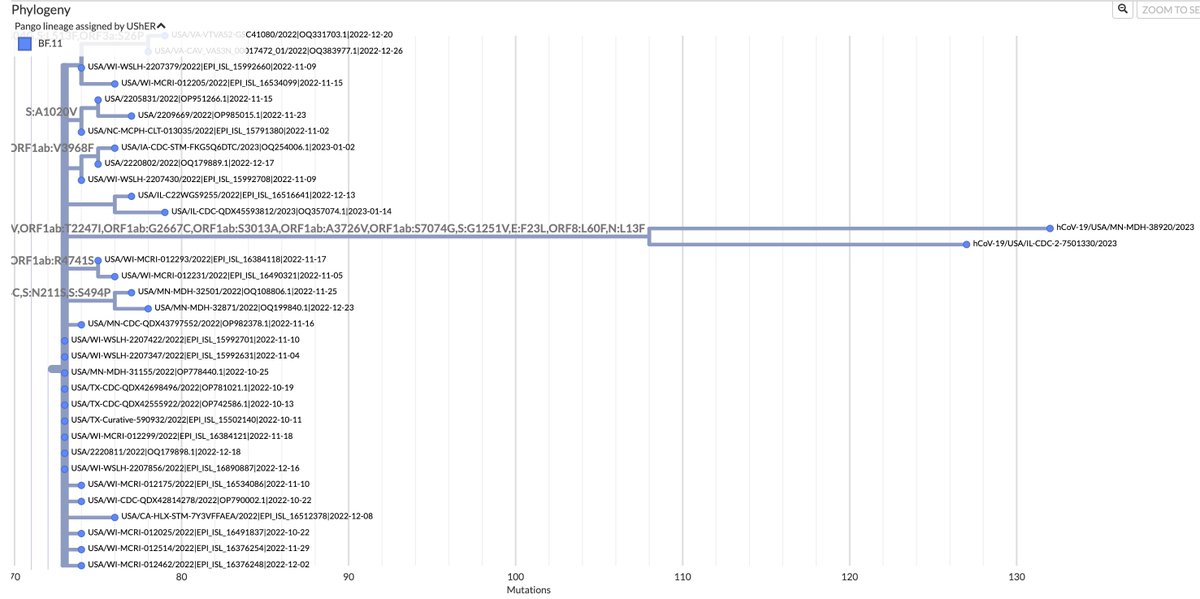
Delta's still hanging around—and packing heat, in the form of 30+ spike mutations. As w/other recent Delta singlets, this is very likely 1 chronically infected person‚ not a circulating variant. But it's a healthy reminder that the prospect of a Delta comeback always looms. 1/15 
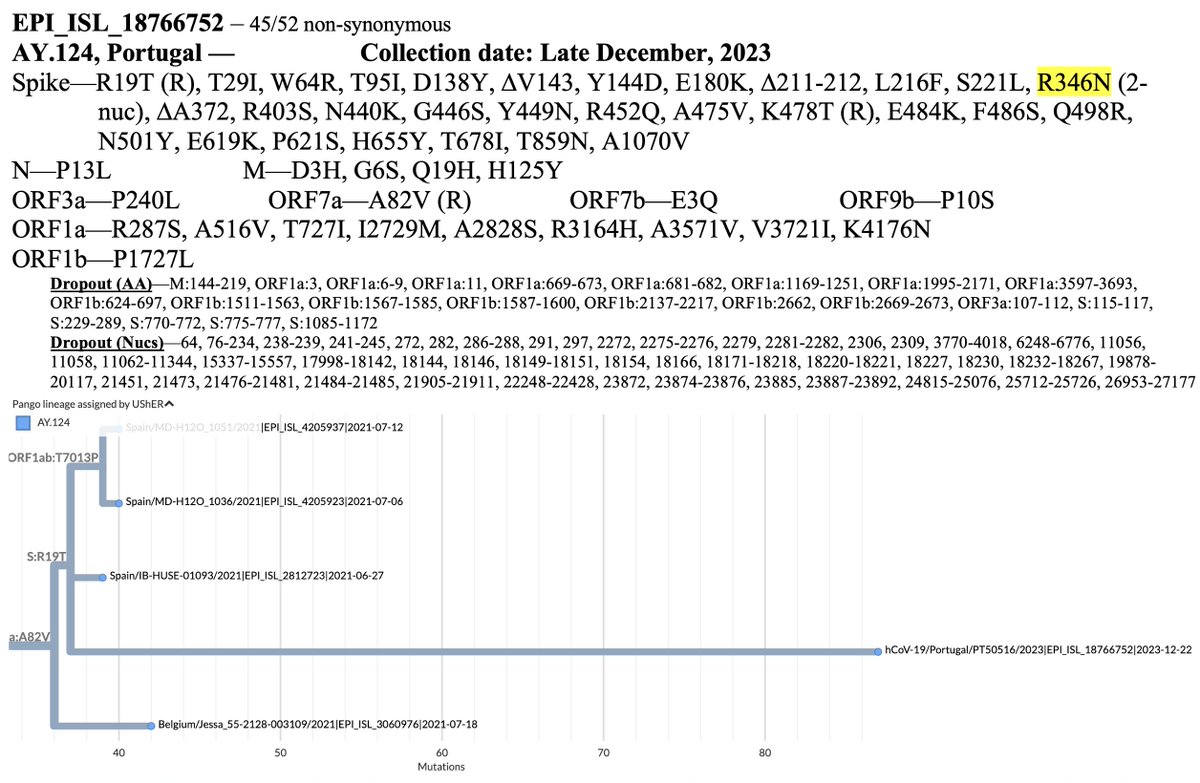
A few aspects of this Delta worth noting:
• About 9% of the genome lacks coverage, including >10% of spike, so the mutations indicated are an undercount. Hopefully, we never see this particular virus again and will never know what's hiding behind the dropout. 2/15
• About 9% of the genome lacks coverage, including >10% of spike, so the mutations indicated are an undercount. Hopefully, we never see this particular virus again and will never know what's hiding behind the dropout. 2/15
• 45/52 nucleotide mutations are non-synonymous (amino-acid changing). This indicates positive selection—i.e. selection for advantageous mutations. Such a high non-synonymous % is very typical of chronic infections, including the OG Omicron. 3/15
https://twitter.com/sergeilkp/status/1465134289603858435
• S:K478T reversion + R452Q
What's notable here is that, as @Asinickle1 has pointed out, K478T and mutation away from R452 also characterized the other recent extremely mutated Delta (described previously in thread below). 4/15
What's notable here is that, as @Asinickle1 has pointed out, K478T and mutation away from R452 also characterized the other recent extremely mutated Delta (described previously in thread below). 4/15
https://twitter.com/LongDesertTrain/status/1681671270377701380
• S:Q498R-N501Y
This is another similarity to the Indonesian Delta referenced above. Q498R reduces ACE2 binding by itself, but when combined with N501Y, it causes an 387-fold increase in ACE2 affinity. See @jbloom_lab, @tylernstarr, @veeslerlab, @Dr_MattMcCallum paper 5/15
This is another similarity to the Indonesian Delta referenced above. Q498R reduces ACE2 binding by itself, but when combined with N501Y, it causes an 387-fold increase in ACE2 affinity. See @jbloom_lab, @tylernstarr, @veeslerlab, @Dr_MattMcCallum paper 5/15

These are the only Deltas ever to have Q498R-N501Y. They also have far more spike mutations than any other Deltas. That's not a coincidence. The ACE2-binding bump provides space for antibody-evading RBD mutations, most of which reduce ACE2 affinity. 6/15
https://twitter.com/LongDesertTrain/status/1562183568389988353
• S:R346N
This is an extremely rare 2-nucleotide mutation at one of the most antigenically important AA sites in spike.
• S:R403S
Another extremely rare mutation at an important spike location that is almost exclusively seen in chronic-infection sequences. 7/15
This is an extremely rare 2-nucleotide mutation at one of the most antigenically important AA sites in spike.
• S:R403S
Another extremely rare mutation at an important spike location that is almost exclusively seen in chronic-infection sequences. 7/15

• Glycan #1
A glycan is basically a sugar attached to the outside of spike. With the T19R reversion, this Delta regains the N17 glycan that was originally present in SARS-CoV-2 (and subsequently lost in Omicron due to the T19I mutation). h/t @Sinickle on this one. 8/15
A glycan is basically a sugar attached to the outside of spike. With the T19R reversion, this Delta regains the N17 glycan that was originally present in SARS-CoV-2 (and subsequently lost in Omicron due to the T19I mutation). h/t @Sinickle on this one. 8/15

• Glycan #2
Glycans can have many functions—as glycan experts like @FabrizioChiodo can explain—but when they occur in the receptor binding domain (RBD), there's a good chance they function to shield exposed sites from antibodies. See @PaulBieniasz 9/15

Glycans can have many functions—as glycan experts like @FabrizioChiodo can explain—but when they occur in the receptor binding domain (RBD), there's a good chance they function to shield exposed sites from antibodies. See @PaulBieniasz 9/15
https://twitter.com/PaulBieniasz/status/1744727542303789096

• Glycan #2 cont.
In this Delta, the deletion of A372 adds a glycan at N370. The formula for a glycan is N-(not P)-S/T, and with ∆372, S:370-371-373 becomes N-S-S.
Not every N-(not P)-S/T motif adds a glycan. But in this case, we know a glycan results. How do we know? 10/15
In this Delta, the deletion of A372 adds a glycan at N370. The formula for a glycan is N-(not P)-S/T, and with ∆372, S:370-371-373 becomes N-S-S.
Not every N-(not P)-S/T motif adds a glycan. But in this case, we know a glycan results. How do we know? 10/15

Because SARS-CoV-1 & all related Bat-CoVs have A372T, which forms an important glycan. It likely influences the open/closed state of the RBD, which in turn affects ACE2 affinity and immune evasion. One study found that A372T greatly reduced infectivity in human lung cells. 11/15




• ORF7a:A82V Reversion
This is *by far* the most common mutation in chronic-infection Delta sequences. It seems to be in about half of them. This seems hard to square with the frequent large ORF7a deletions, stop codons, & TRS-destroying mutations that have occurred... 12/15
This is *by far* the most common mutation in chronic-infection Delta sequences. It seems to be in about half of them. This seems hard to square with the frequent large ORF7a deletions, stop codons, & TRS-destroying mutations that have occurred... 12/15
…frequently over the last few years. But there are several other ORF7a mutations convergent in chronics (E22D, N43X, S81P, L96P, A105V), so it clearly is doing something in these patients. What precisely is ORF7a:A82V doing? I have no idea. Maybe someone else does?
13/15
13/15
It seems likely that in some genetic & individual contexts ORF7a is disposable, while in other contexts, it performs a vital function, likely involving immune evasion. The presence or absence of ORF6:D61L is one factor that clearly influences ORF7a. 14/15
https://twitter.com/LongDesertTrain/status/1718769995830755719
I'll end this thread with a thank you to the work of labs & sequencers all over the world, in this case the lab of @borges__vitor. Without them we would be in the dark.
15/15
15/15
• • •
Missing some Tweet in this thread? You can try to
force a refresh