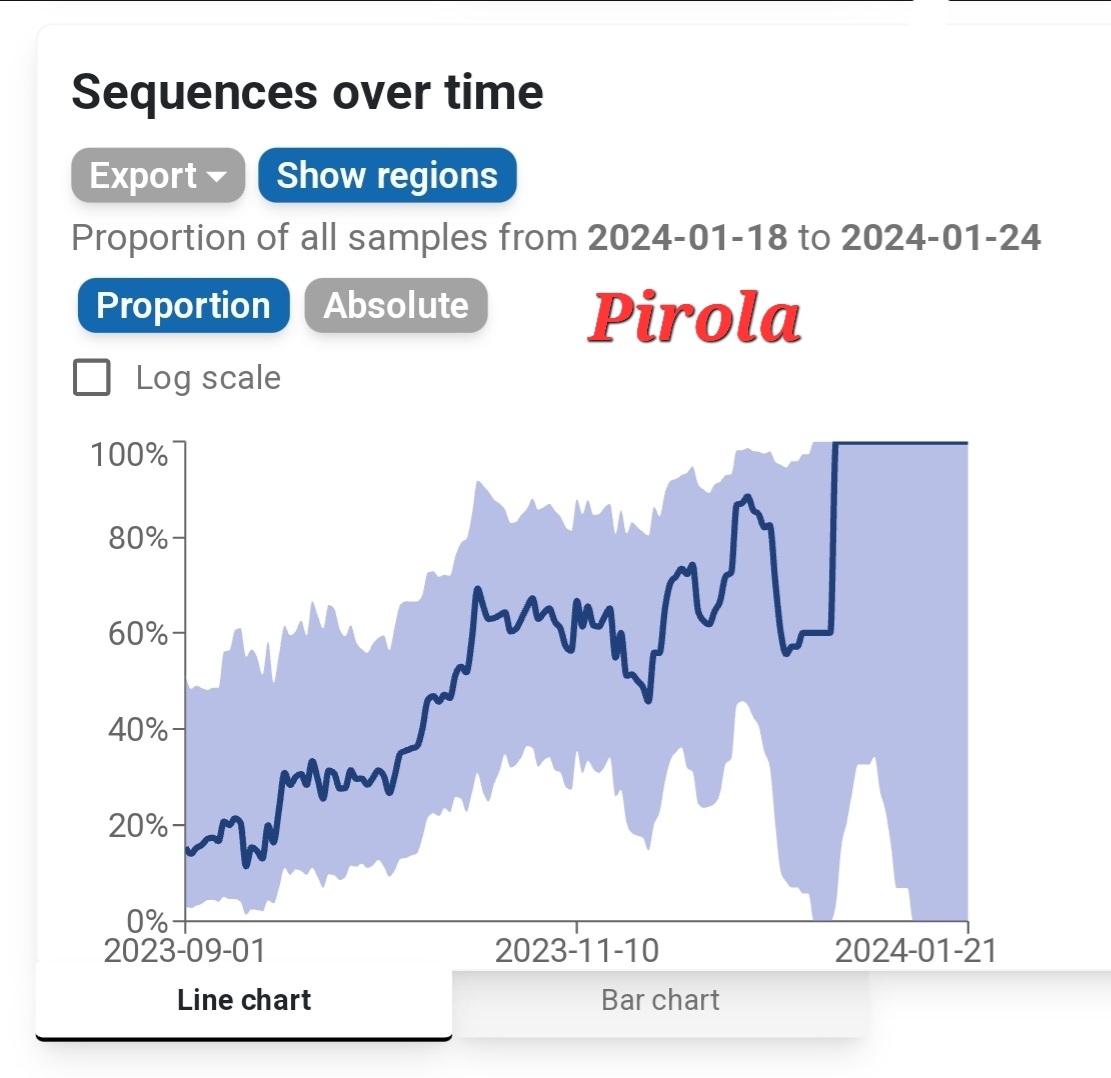
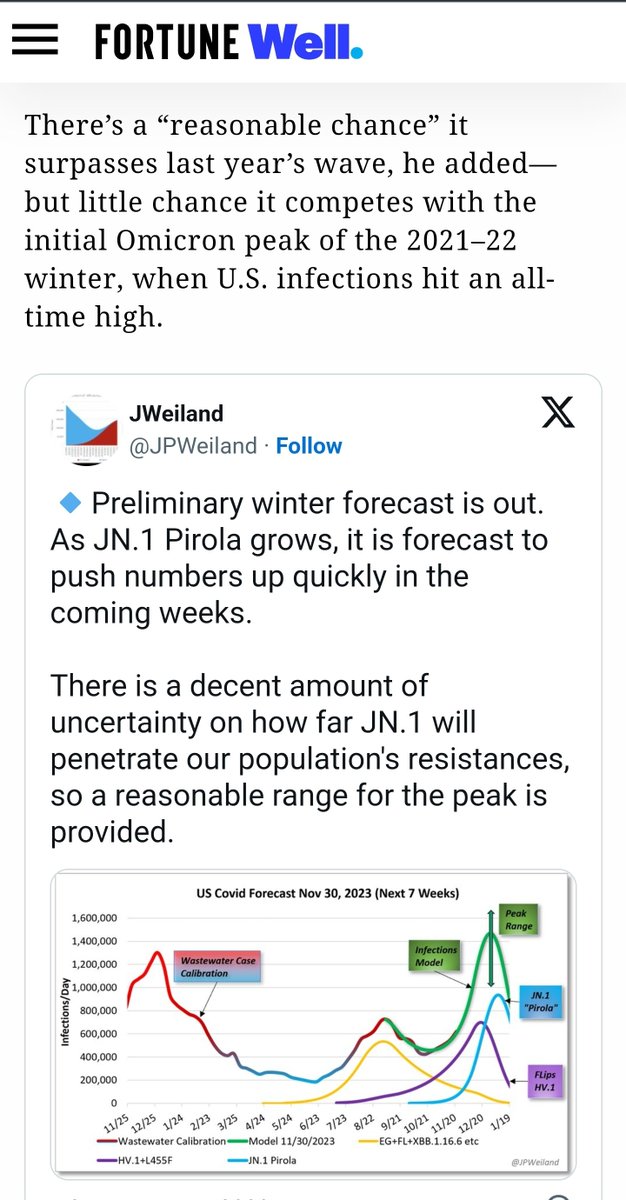
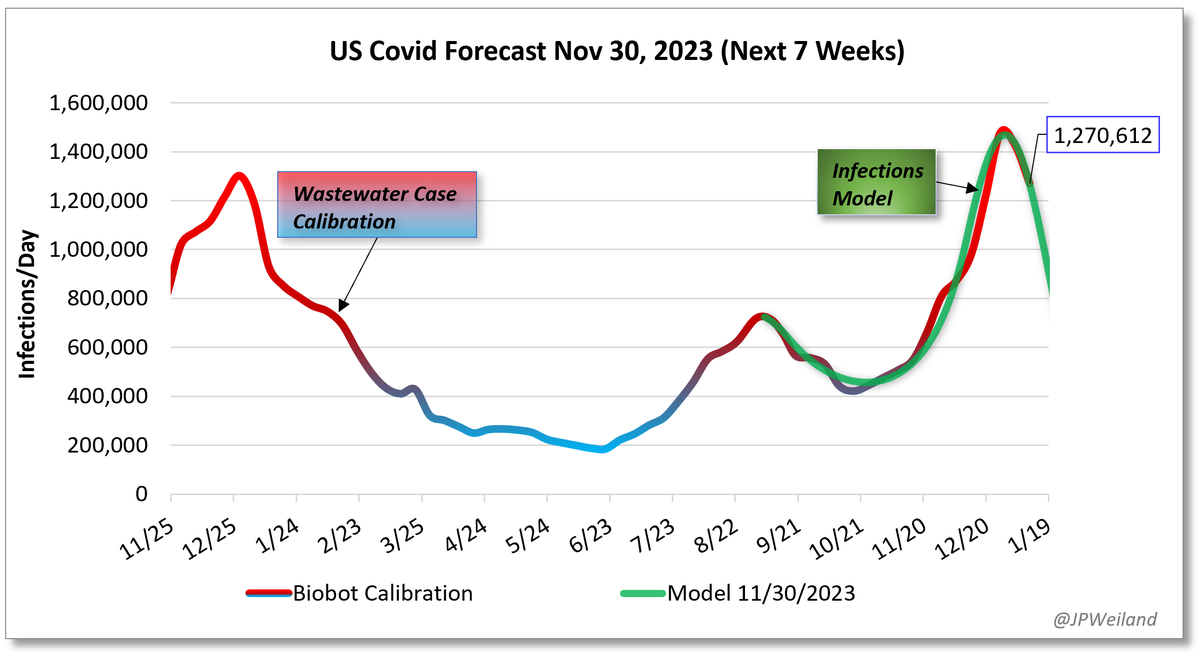
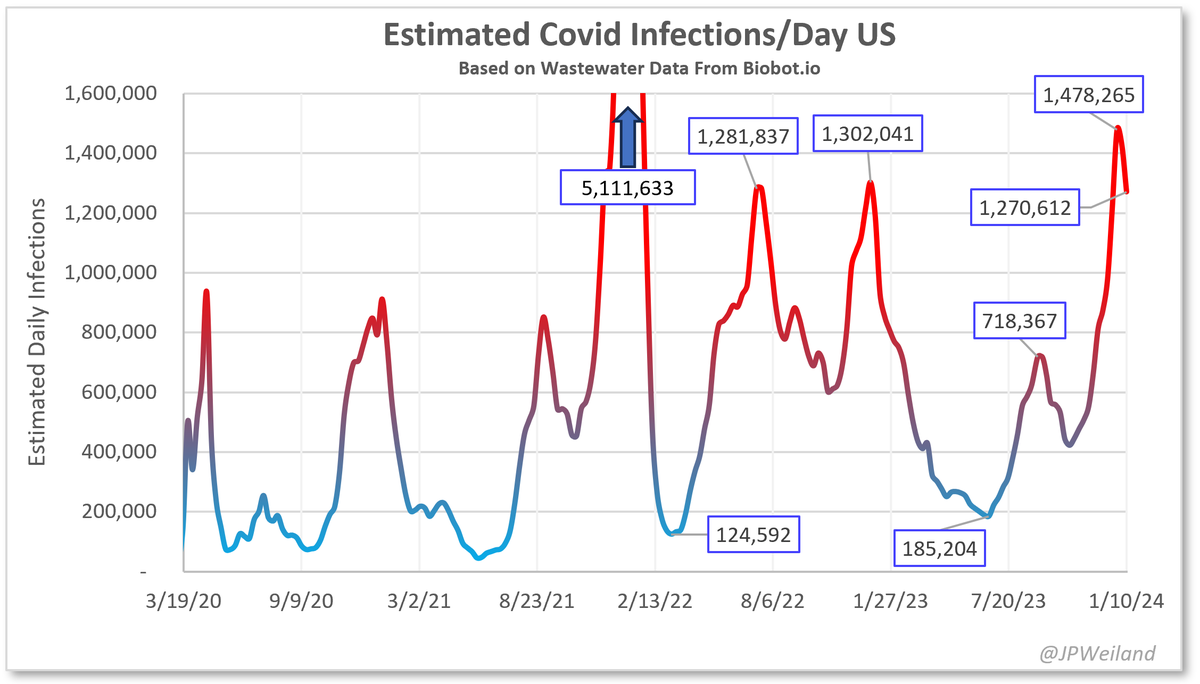
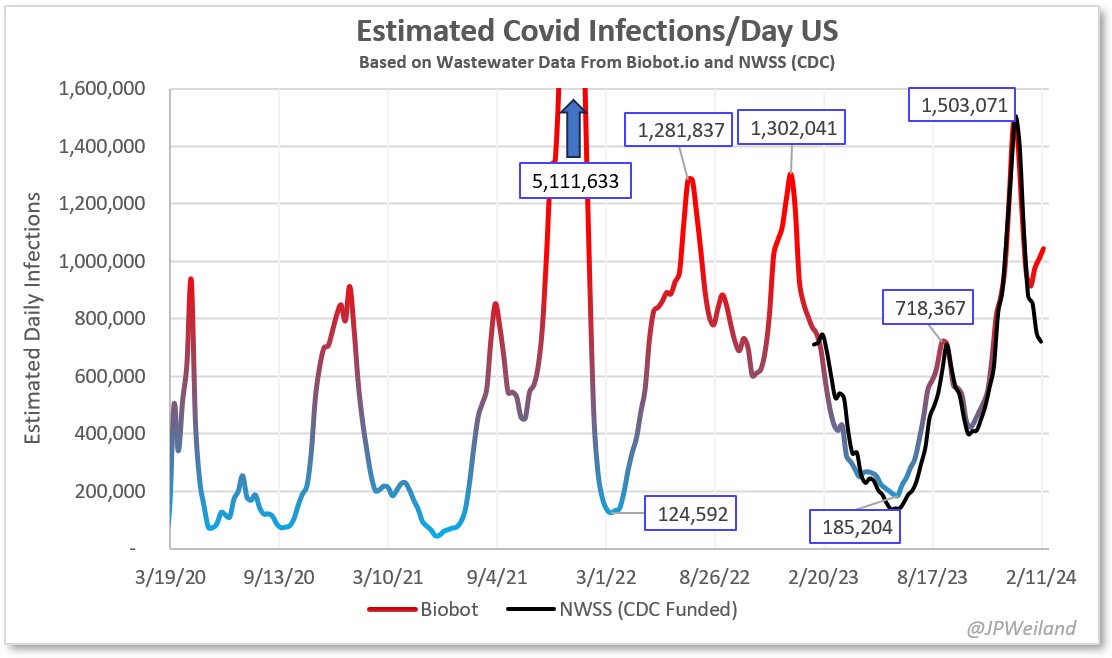
The divergence between Biobot and NWSS continues. NWSS at this point commands a far larger sampling pool than Biobot. Therefore, if we weight toward NWSS data:
🔸780,000 new infections/day
🔸1 in every 430 became infected today
🔸1 in every 43 people currently infected
🔸780,000 new infections/day
🔸1 in every 430 became infected today
🔸1 in every 43 people currently infected
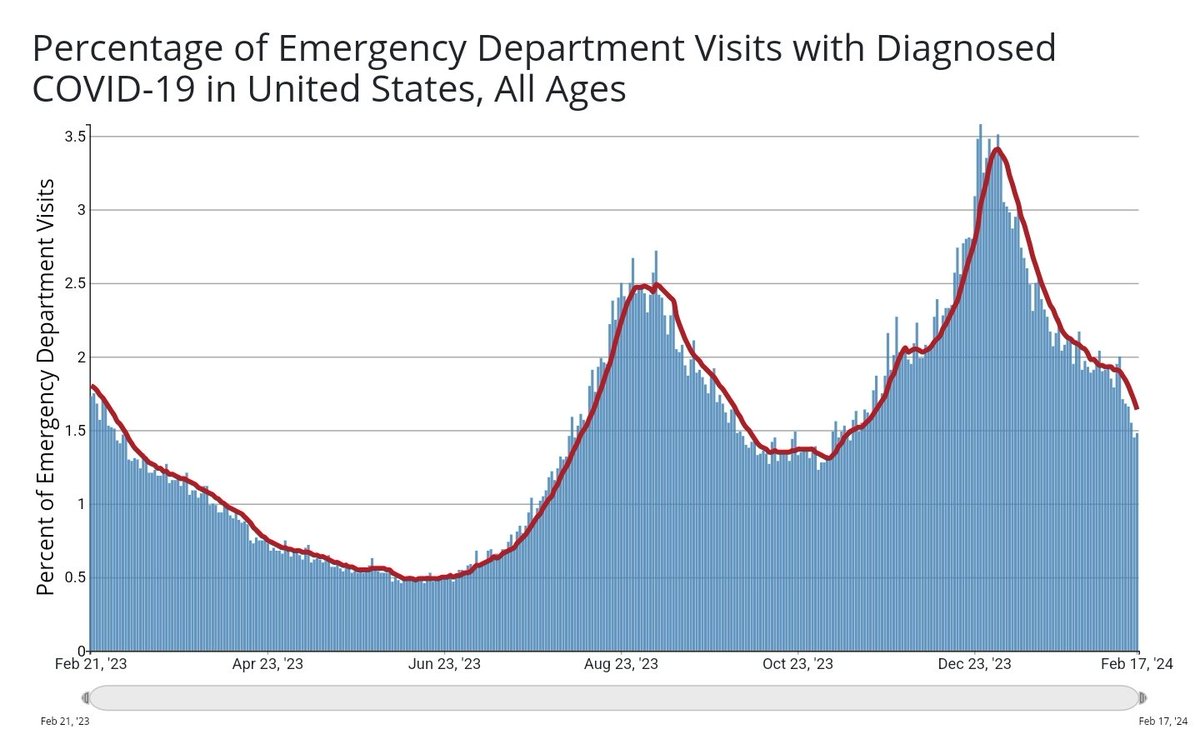
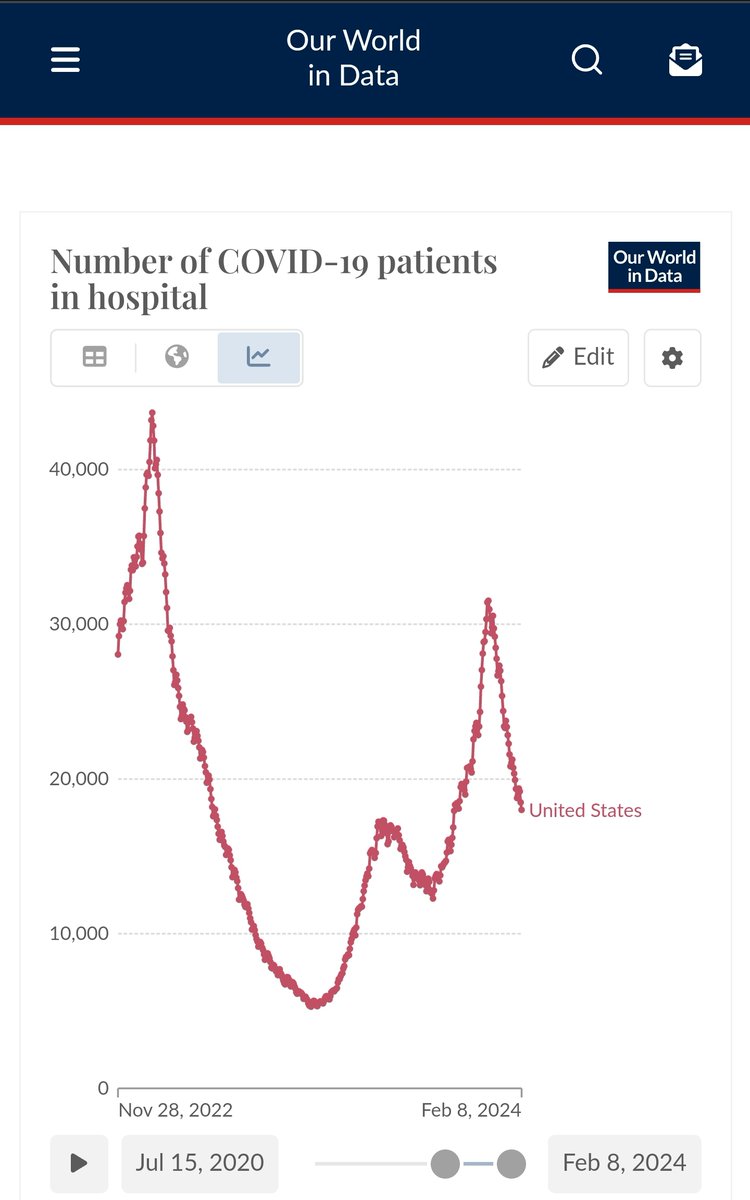
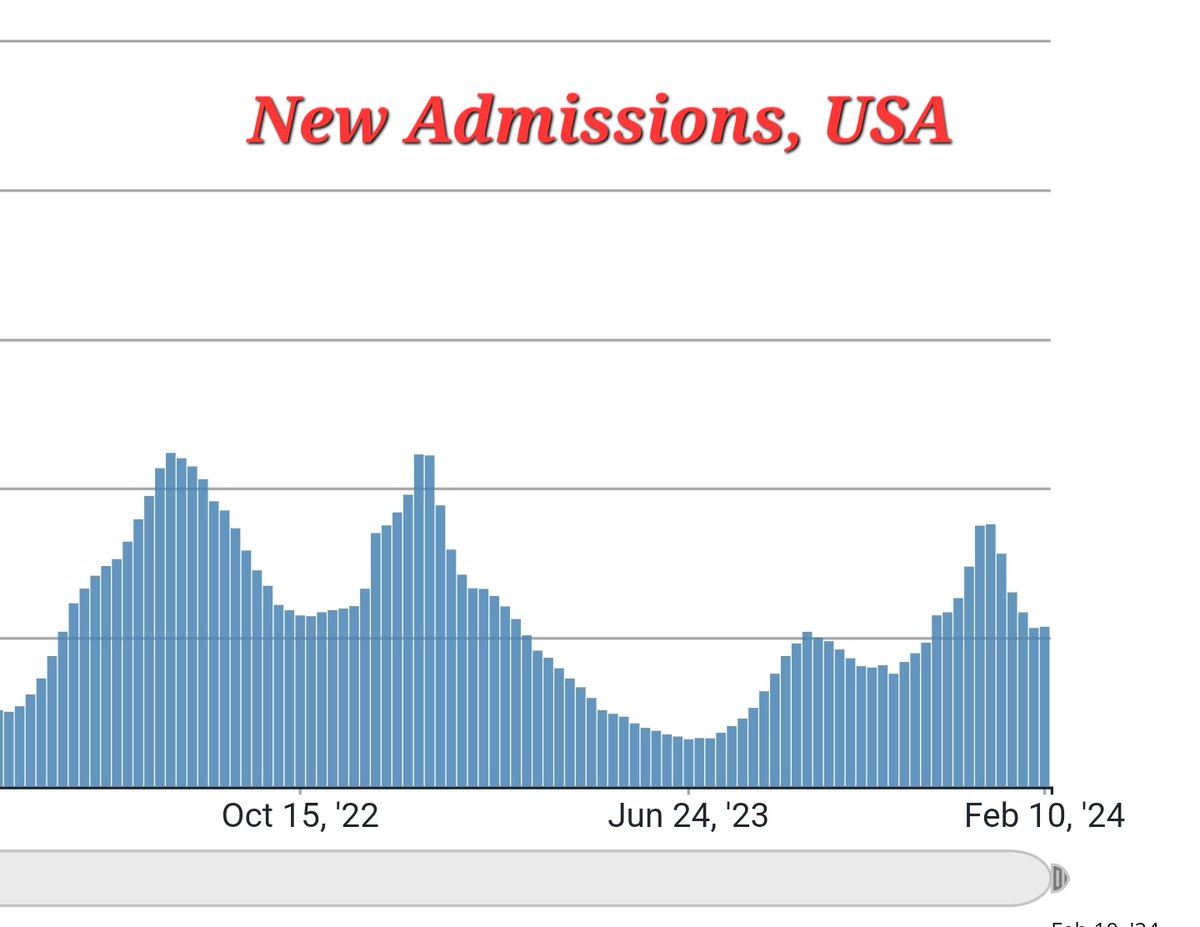
% of ED visits and # hospitalized track NWSS well, but new admissions was flat last week (still much closer to NWSS). Normally all of these data sets have good agreement, but not today! Makes my job more difficult, for sure.



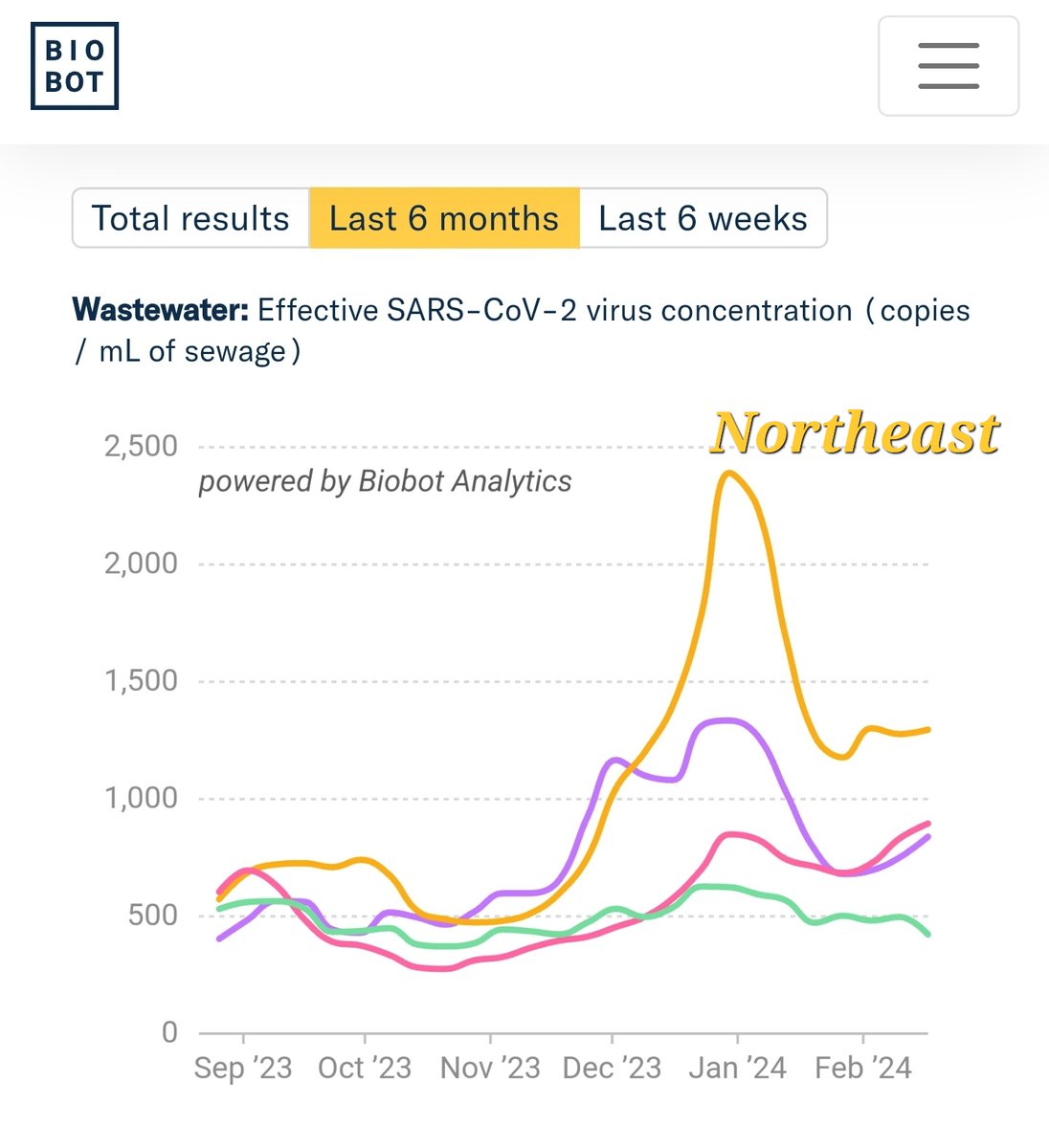
Biobot is the most anomalous in the Northeast. I absolutely don't believe that levels are highest there. Also I think they undersample the South.
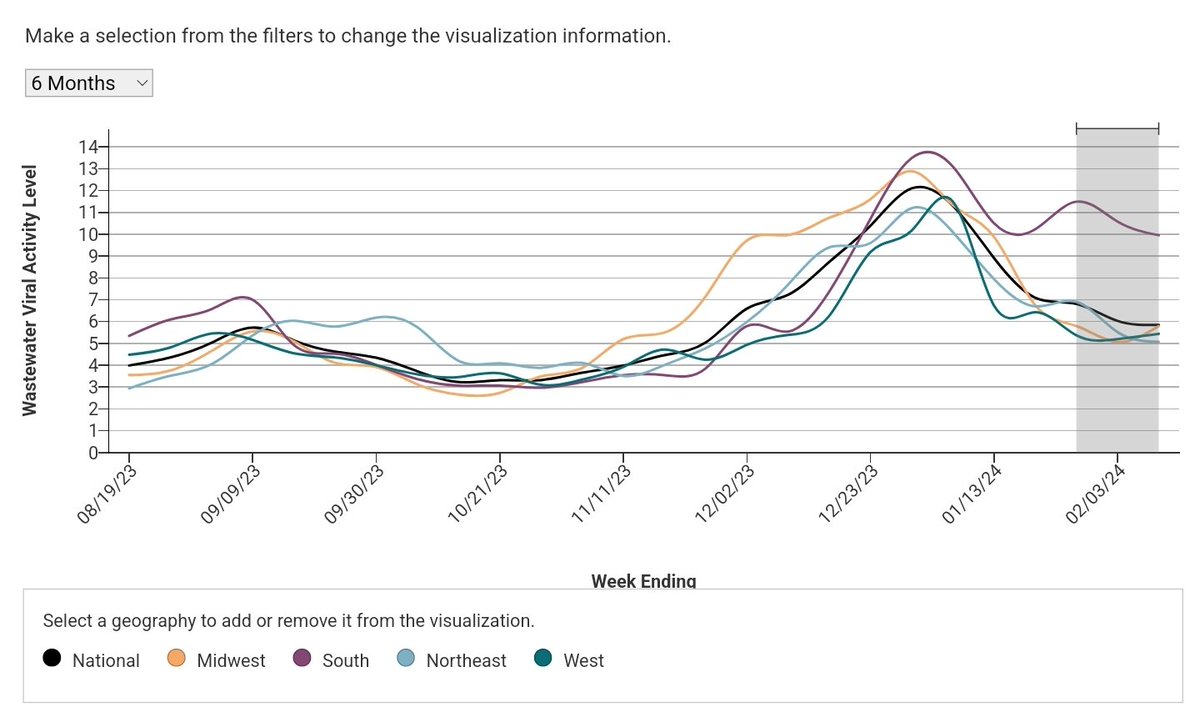
NWSS and hosp metrics still show very high levels in the deep south. I'm starting to trust NWSS data more.


NWSS and hosp metrics still show very high levels in the deep south. I'm starting to trust NWSS data more.
• • •
Missing some Tweet in this thread? You can try to
force a refresh