Calling all genomics Jocks.
We sequenced vax treated OvCar3 cell lines that were washed and passaged such that the vaccine was diluted out and only cells with DNA inside are present.
Our collaborators who did this work also stained for spike expression.
We sequenced vax treated OvCar3 cell lines that were washed and passaged such that the vaccine was diluted out and only cells with DNA inside are present.
Our collaborators who did this work also stained for spike expression.
60% of cells are positive for spike with IHC
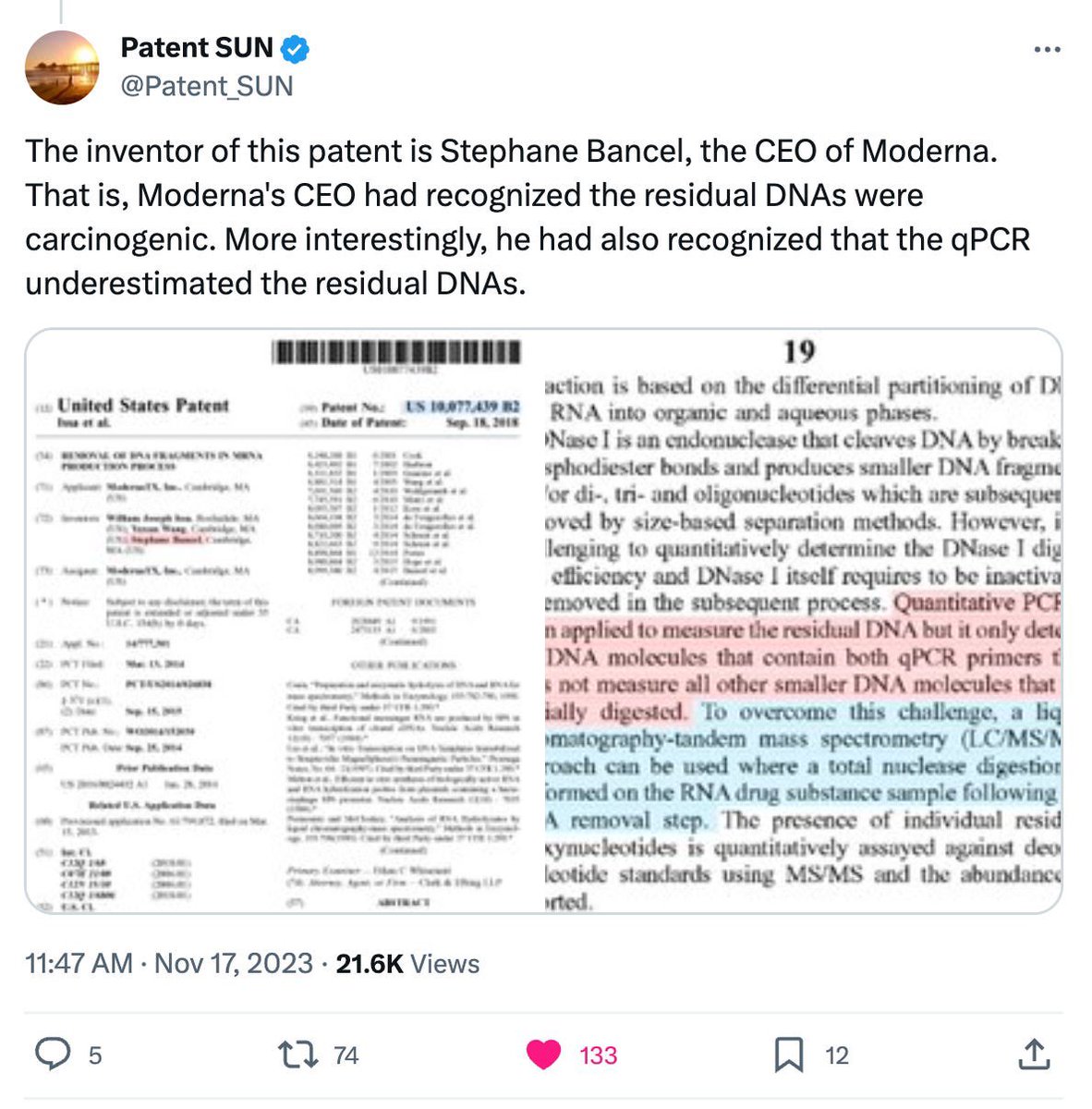
We qPCRd these cells for spike,SV40 and Ori in bulk.
Not single cell measurement.
Positive with CTs similar to human RNAP.
We performed whole genome sequencing.
The entire vax plasmid can be reassembled at 3,000X coverage (Top Track).
We qPCRd these cells for spike,SV40 and Ori in bulk.
Not single cell measurement.
Positive with CTs similar to human RNAP.
We performed whole genome sequencing.
The entire vax plasmid can be reassembled at 3,000X coverage (Top Track).

This is at 30X coverage of the human genome so 100x higher coverage for the plasmid than the human genome?
As a control we also sequenced the vax alone.
It’s at 44,000X coverage.
You can see the left side of the IGV view has no SNPs. That’s the vector.
The spike has SNPs
As a control we also sequenced the vax alone.
It’s at 44,000X coverage.
You can see the left side of the IGV view has no SNPs. That’s the vector.
The spike has SNPs
The SNPs in the plasmid are in this hairpin loop. This is the F1 Ori responsible for making ssDNA from the plasmid/phagemid. 

Image courtesy of Stephen McLaughlin via RNA fold.
A close up on the allele frequencies of these events.
Notice, they are 3 variants in tandem and in phase.
Right in the hairpin loop.
Notice, they are 3 variants in tandem and in phase.
Right in the hairpin loop.

The question for the genome jocks…
Would you expect at 30X coverage of the human genome that you would get
3X coverage of the vax
30X coverage
300X
Or
3,000x coverage.
I wasn’t expecting 3000X coverage.
Would you expect at 30X coverage of the human genome that you would get
3X coverage of the vax
30X coverage
300X
Or
3,000x coverage.
I wasn’t expecting 3000X coverage.
For reference…
depth of coverage in sequencing should reflect a ratio of the DNA in the cell.
For instance, when you sequence human cells you usually get 100-500X coverage of the Mito for every 1X of the human genome.
Higher coverage for Mito in muscle cells.
Similar coverage differences are seen with cholorlast genomes in plant cells because these organelle genomes are multi copy per cell.
Plasmids are usually in cells at 100 copies per genomic equivalents.
How is the vax getting to these copy numbers if it’s not replication competent?
depth of coverage in sequencing should reflect a ratio of the DNA in the cell.
For instance, when you sequence human cells you usually get 100-500X coverage of the Mito for every 1X of the human genome.
Higher coverage for Mito in muscle cells.
Similar coverage differences are seen with cholorlast genomes in plant cells because these organelle genomes are multi copy per cell.
Plasmids are usually in cells at 100 copies per genomic equivalents.
How is the vax getting to these copy numbers if it’s not replication competent?
It could be a very high MOI (multiplicity of infection).
Fancy term for lots of LNPs per cell.
But I can’t imagine you can get 100 LNPs per cell without killing the cell.
Fancy term for lots of LNPs per cell.
But I can’t imagine you can get 100 LNPs per cell without killing the cell.
So 4 SNPs in the plasmid vector which are not seen in the vaccine plasmids that don’t touch OvCar3 and they are both origins of replication.
And they have coverage peaks.
That’s implies the DNA is being replicated by OvCar3 and making replication errors.
But..
And they have coverage peaks.
That’s implies the DNA is being replicated by OvCar3 and making replication errors.
But..

Since the fragments are short the replication does not go very far and makes narrow coverage peaks.
The SV40 origin SNP is in this stem.
That’s strong evidence that the DNA is in a cell that’s modifying it and partial replication is occurring.
The SV40 origin SNP is in this stem.
That’s strong evidence that the DNA is in a cell that’s modifying it and partial replication is occurring.

A strong argument for removing all origins of replication from vaccines as small amounts of contamination can lead to more once in a cell.
@Shaikan100 Just wasn’t expecting any in the plasmid backbone as we don’t see that when we sequence the vax directly.
Something about transfecting it into cell lines created those variants in the plasmid.
Something about transfecting it into cell lines created those variants in the plasmid.
@Waldrada So 600X for the vax and 30X for the genomic DNA.
@KateYandell care correct your statements regarding how this DNA doesn’t get into the cell?
@KateYandell care correct your statements regarding how this DNA doesn’t get into the cell?
• • •
Missing some Tweet in this thread? You can try to
force a refresh