How common are SARS-CoV-2 persistent infections (revisited).
Last summer I tried to calculate how frequent persistent SARS-CoV-2 infections were from sequence info.
Since there was a recent paper on this, I thought I would revisit my calculations.
1/
Last summer I tried to calculate how frequent persistent SARS-CoV-2 infections were from sequence info.
Since there was a recent paper on this, I thought I would revisit my calculations.
1/
https://twitter.com/SolidEvidence/status/1674538153347276801
My calculations depend on 2 assumptions.
1. Sequences with Orf1a:K1795Q usually (>80% of the time) come from persistent infections (excluding Gamma and BS.1).
2. About 4-10% of all long persistent infections acquire Orf1a:K1795Q.
2/
1. Sequences with Orf1a:K1795Q usually (>80% of the time) come from persistent infections (excluding Gamma and BS.1).
2. About 4-10% of all long persistent infections acquire Orf1a:K1795Q.
2/
Let's revisit the first assumption with new data.
How many times did Orf1a:K1795Q appear in sequences in the last 6 months?
69
How many of those were ‘older’ lineages?
About 60.
So definitely over 80%. Assumption still stands.
3/
How many times did Orf1a:K1795Q appear in sequences in the last 6 months?
69
How many of those were ‘older’ lineages?
About 60.
So definitely over 80%. Assumption still stands.
3/

Now the second assumption.
How many times has Orf1a:1795Q appeared in BA.4* lineages since it stopped circulating (which I set at 2/1/23)?
21
How many total BA.4* sequences were reported during that time?
204
21/204 = 10.3%. About the same. Assumption still stands.
4/
How many times has Orf1a:1795Q appeared in BA.4* lineages since it stopped circulating (which I set at 2/1/23)?
21
How many total BA.4* sequences were reported during that time?
204
21/204 = 10.3%. About the same. Assumption still stands.
4/

So if the ~60 persistent infections with Orf1a:1795Q reported in the last 6 months represent 5-10%, that means the actual number should have been 600-1200 out of 479k total sequences.
= 0.125-0.25% of all sequences. Similar number as before.
5/
= 0.125-0.25% of all sequences. Similar number as before.
5/
This is a bit lower range than what was estimated in the paper. They estimated 0.1-0.5% were more that 60 days, but I suspect what I am measuring is actually a smaller cohort of people with longer infections.
6/
nature.com/articles/s4158…
6/
nature.com/articles/s4158…
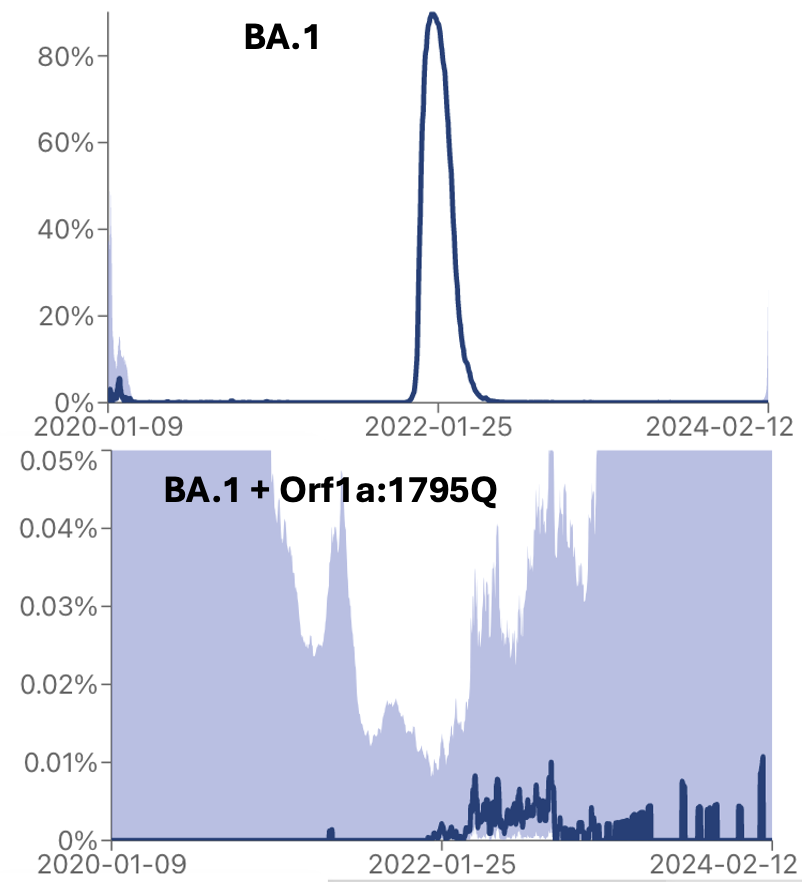
I was really conservative where I drew the line for when lineages stopped circulating, so for my calculation the infection really had to have lasted at least 4 months.
Look at the vasty different distribution of seqs of BA.1* and BA.1*+1795Q.
7/
Look at the vasty different distribution of seqs of BA.1* and BA.1*+1795Q.
7/
Interestingly, none of the patients in the Nature paper had actually picked up Orf1a:K1795Q. I think this is because these patients had not ‘incubated’ long enough.
They did identify two mutations that did appear somewhat ‘persistent specific’: Orf1a:1638I and 4311I.
8/
They did identify two mutations that did appear somewhat ‘persistent specific’: Orf1a:1638I and 4311I.
8/
Orf1a:1638I is clearly highly prevalent in persistent infections. Look at the difference between BA.1* and BA.1*+1638I.
Not as selective as 1795Q, but clearly linked to persistent infection.
9/
Not as selective as 1795Q, but clearly linked to persistent infection.
9/

Orf1a:4311I was not as obvious, but it did still show a small degree of enrichment in persistent infections.
10/
10/

So the summary really hasn’t changed since last summer.
At least 1 in 1000 SARS-CoV-2 sequences appear be derived from very long persistent infections.
11/
At least 1 in 1000 SARS-CoV-2 sequences appear be derived from very long persistent infections.
11/
Caveats:
-The data isn’t perfect. At least some of the samples had the wrong date entered, and at least some of the samples represent patients that were sampled multiple times.
-We don’t know whether people with persistent infections are more or less likely to be sampled.
12/
-The data isn’t perfect. At least some of the samples had the wrong date entered, and at least some of the samples represent patients that were sampled multiple times.
-We don’t know whether people with persistent infections are more or less likely to be sampled.
12/
Finally, the most important caveat (also pointed out by @LongDesertTrain). The seqs are all from nasal samples.
How many more persistent infections occur with the virus replicating elsewhere in the body? We know this occurs.
IMO, more research is needed on this.
13/
How many more persistent infections occur with the virus replicating elsewhere in the body? We know this occurs.
IMO, more research is needed on this.
13/
• • •
Missing some Tweet in this thread? You can try to
force a refresh