Some thoughts on the ability to distinguish populations with genetic variation, why that means little for trait differences, and why there are other good reasons to collect diverse data. 🧵
I was pleasantly surprised to see no one mount a strong defense of "biological race" in this thread. Even the people throwing this term around seem to realize it's not supported by data. Instead the conversation shifts to population "distinguishability".
https://twitter.com/SashaGusevPosts/status/1760367948177252797
For example, a random twitterer (left) and a professor (right) emphasizing that genetic variation can be used to "distinguish" populations. And it's true, one can aggregate small per-variant differences into genetic ancestry estimates that often correlate highly with geography.




Moreover, it's true at essentially all scales: ancestry inference in self-reported whites in the US correlates w/ European country references; ancestry inference in self-reported "White British" in the UK correlates with latitude/longitude and counties in the UK; and so on. 
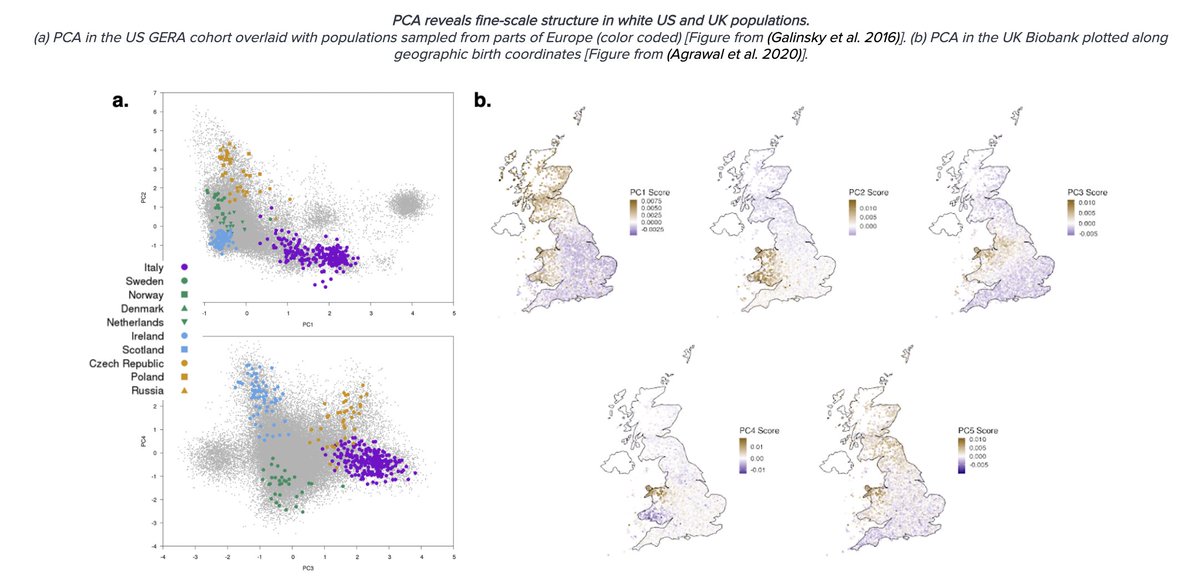
In fact we know, given enough sites, a method like PCA can identify correlations down to a handful of generations (or even pick up families). Of course, no one argues counties/zipcodes are biological units, so "distinguishability" alone is not meaningful. What is meaningful? 
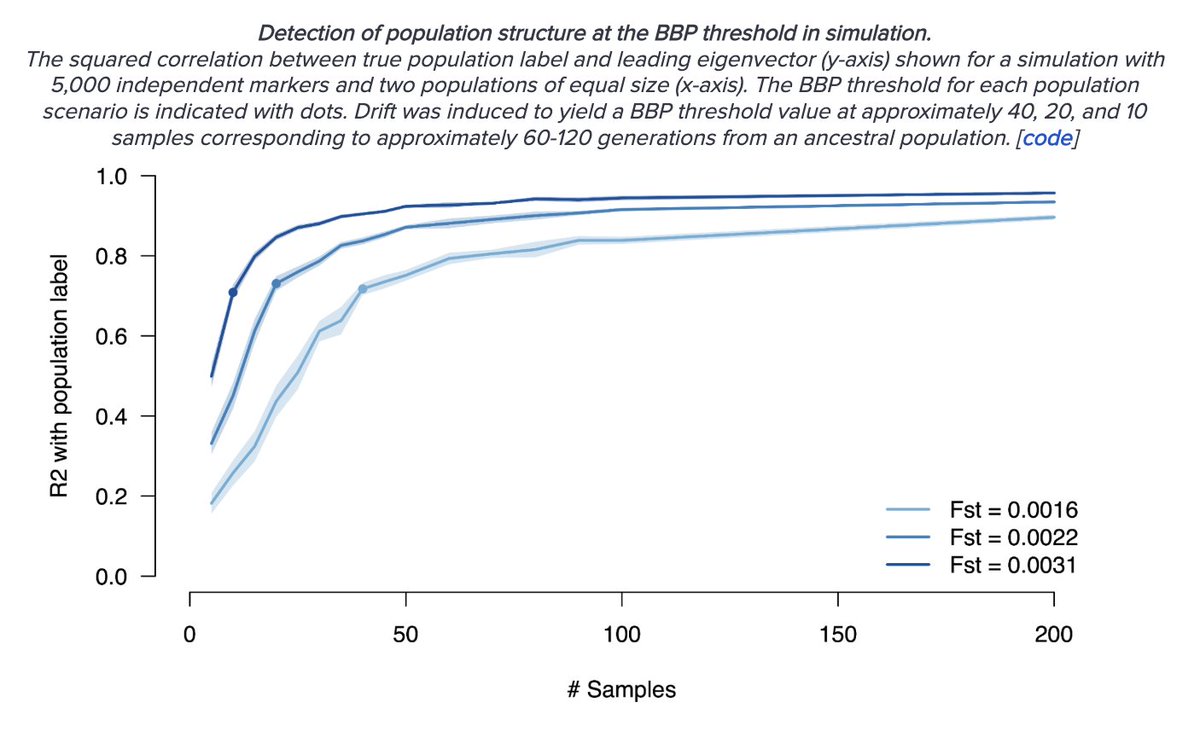
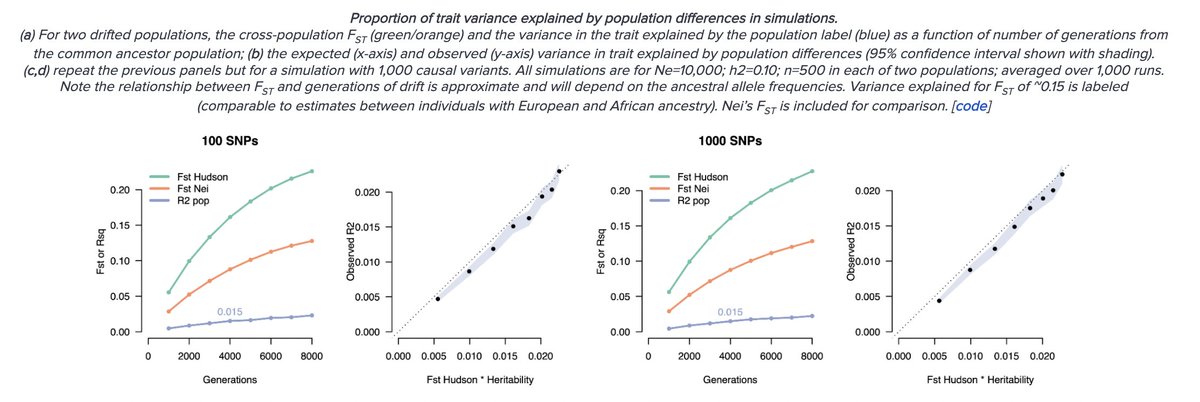
We might be interested in meaningful distinguishability of genetically driven traits. But unlike genetic ancestry, a neutral trait does NOT become more differentiated as you aggregate more variants. So distinguishable ancestry NEED NOT translate into trait differences. 

We even have bounds: the expected between-population neutral trait variance is Fst * heritability. For human populations and traits this is very low (1-8%) even if we take genetic ancestry extremes, and of course these differences are centered at zero and go in either direction. 
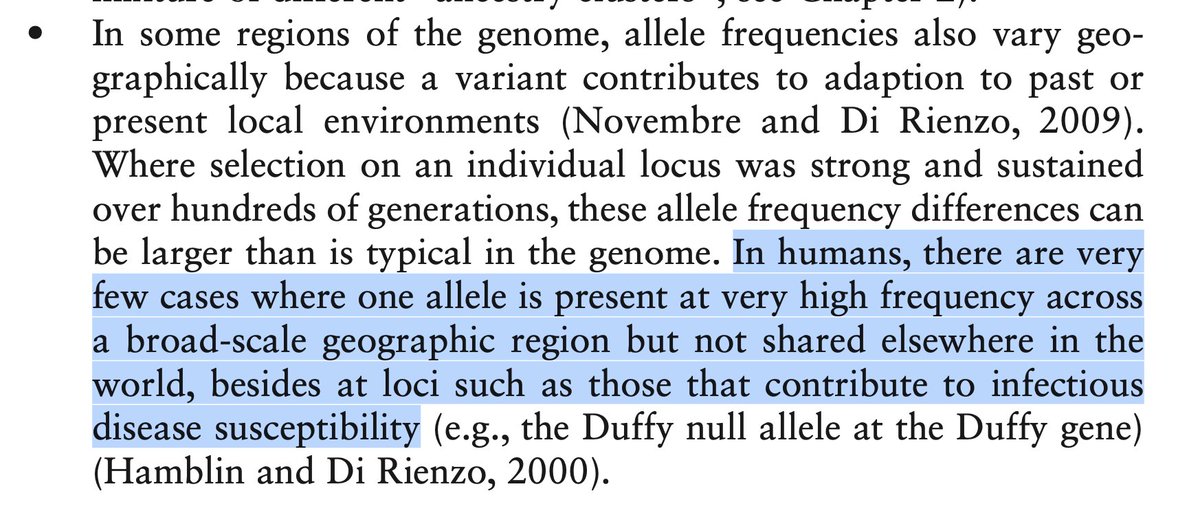
We might be interested in individual large-effect variants with big frequency differences due to bottlenecks (like BRCA) or selective sweeps (like pigment or lactase). In the early genome days there was great speculation that "divergent genes" would explain trait disparities.
Such studies have been run and, as it turns out, "hard sweeps" are very infrequent. This is broadly appreciated in the field but draws intense backlash on twitter, so I'll just quote some sources [ , ] and save the details for later. web.stanford.edu/group/pritchar…
nap.nationalacademies.org/catalog/26902/…


nap.nationalacademies.org/catalog/26902/…

Lastly, perhaps the causal effects of common variants differ substantially between populations (for example due to interactions). Though more work is needed, studies using local ancestry show this does not generally appear to be the case. Details here:
https://twitter.com/SashaGusevPosts/status/1698003409205375329
In short, "distinguishable" ancestry in PCA tells us nothing about traits, either neutral trait means, hard locus-specific selection, or genome-wide effect sizes. So why do we collect diverse data? IMO three good reasons and none of them have to do with trait divergence:
1: Diverse populations are likely to have more diverse *environments*, which (we hope) is useful for understanding the relationships between genetic variation and context [ex: ], as well as enriching for more environmental risk factors.ncbi.nlm.nih.gov/pmc/articles/P…
2: Association studies estimate effects from "tag" SNPs + noise due to LD and frequency. The noise is further amplified across populations leading to poor prediction. Diverse data can improve prediction and increase sensitivity by cleaning up this noise.
https://twitter.com/SashaGusevPosts/status/1724849195117351067
3: Diverse data picks up a few more rare variants (especially non-singletons). These contribute very little to group-specific trait differences, but they can improve imputation, identify novel biology, &be important to their carriers (ex: drug reactions).
https://twitter.com/apragsdale/status/1761927418187554837
TLDR: "distinguishability" is mostly a matter of having enough data points in the analysis. We collect diverse populations not because we expect much trait divergence, but to capture environments, better tag SNPs/LD, and variants in a more useful frequency range. /fin
@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh















