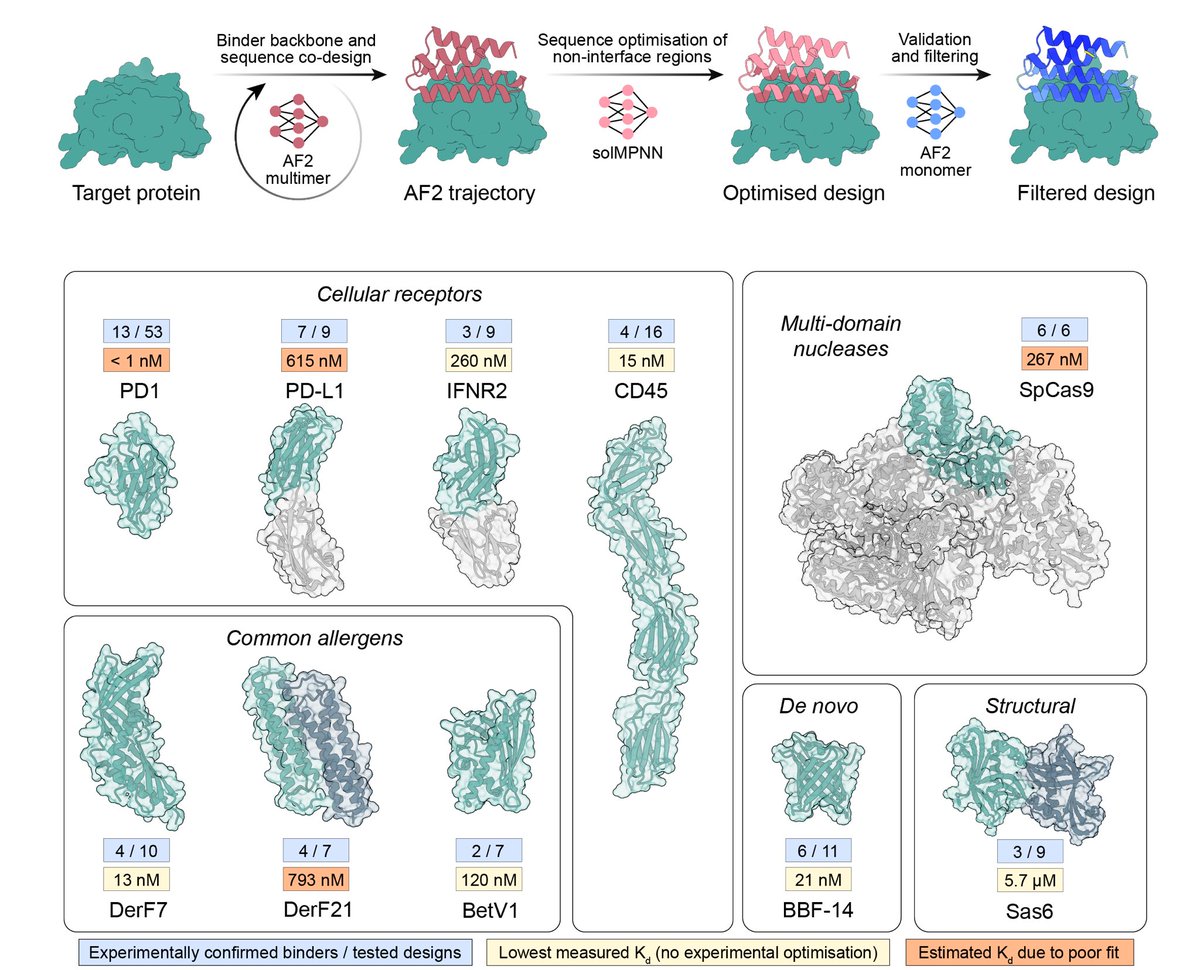
THEY SAID WE COULDN'T DO IT........well, no one actually said that. But in our updated manuscript with @CasperGoverde, @GoldbachNico, and @befcorreia we show you can now design soluble analogues of membrane proteins with preserved functional features!
biorxiv.org/content/10.110…

biorxiv.org/content/10.110…
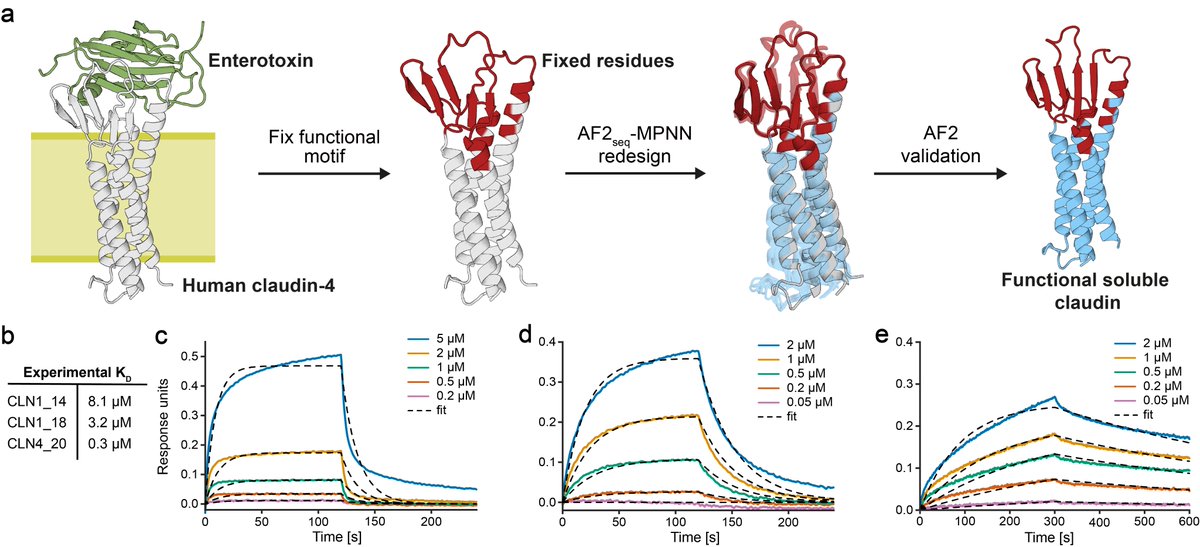
While fixing functional sites during design, we can recapitulate complex interaction sites, such as the Clostridium perfringens enterotoxin binding site on claudins and maintain toxin binding in solution with affinities comparable to its membrane counterpart. 
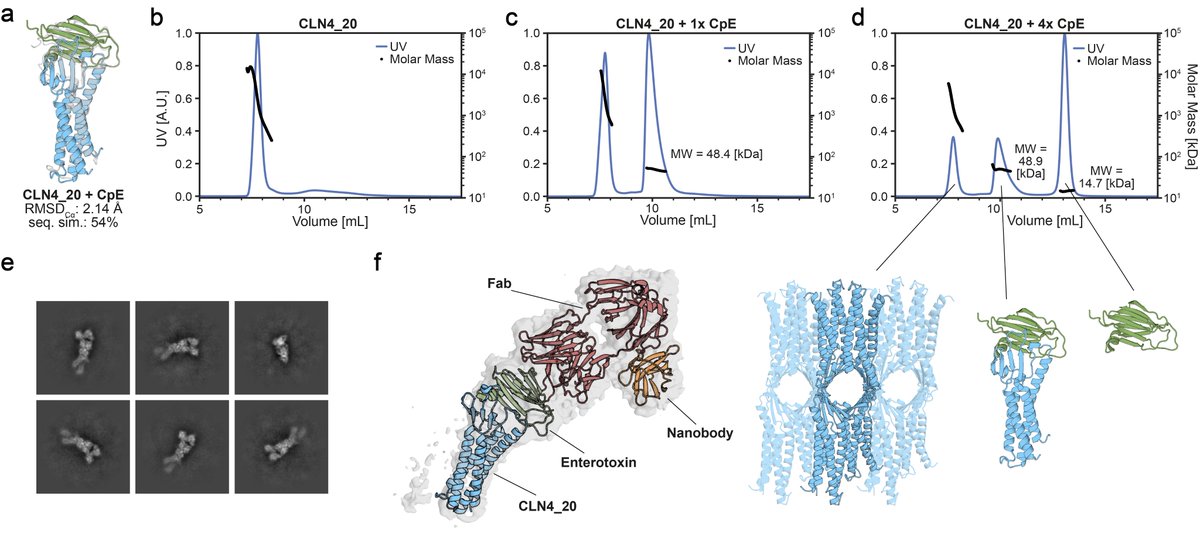
Strikingly, we see that the soluble analogues form high molecular weight species that can be disrupted upon addition of the toxin! Analogous to disassembly of claudin oligomers within tight junctions. @Vecchiology's lab was able to confirm the binding mode using cryoEM. 
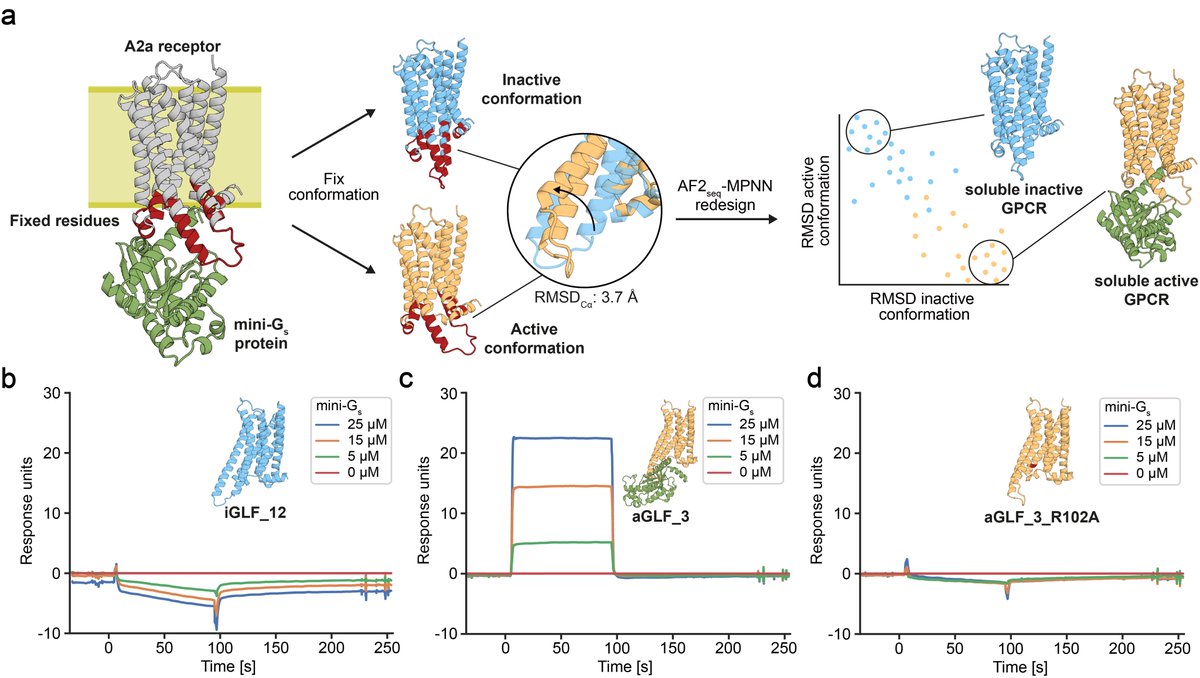
In our previous manuscript version, we have shown that our GPCR analogues can be functionalised using motif grafting approaches... 
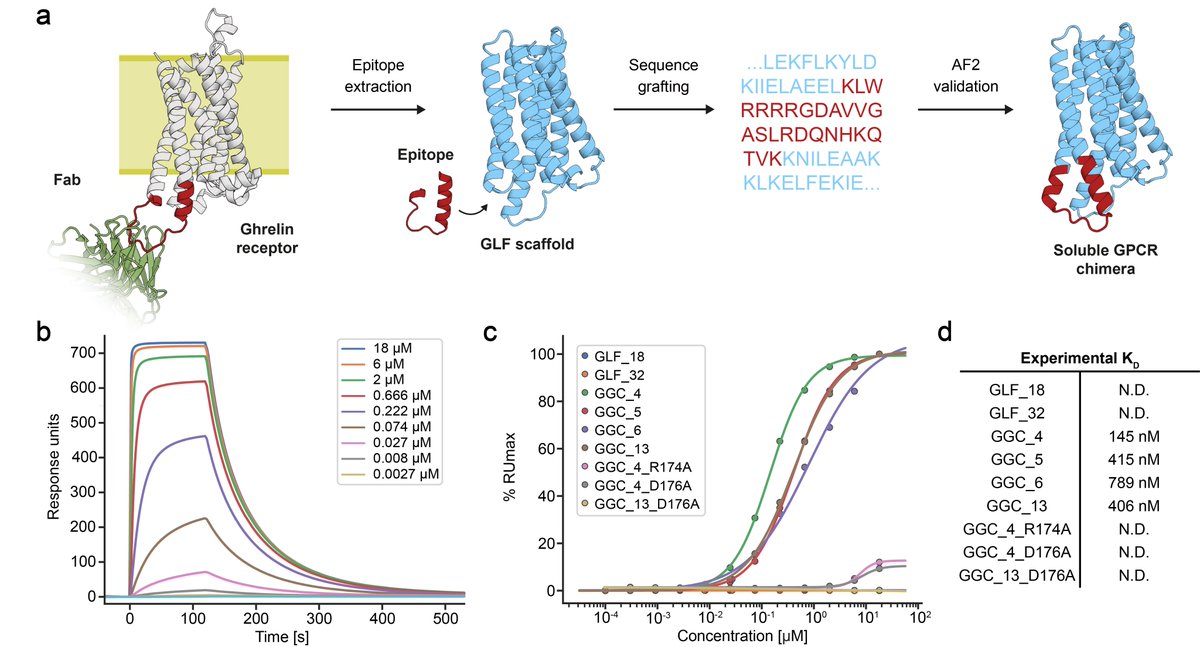
...however, now we show that we can also design GPCR analogues in a conformation-specific way! So we designed them in an active or inactive state and show they can selectively bind G-proteins in solution!!! 
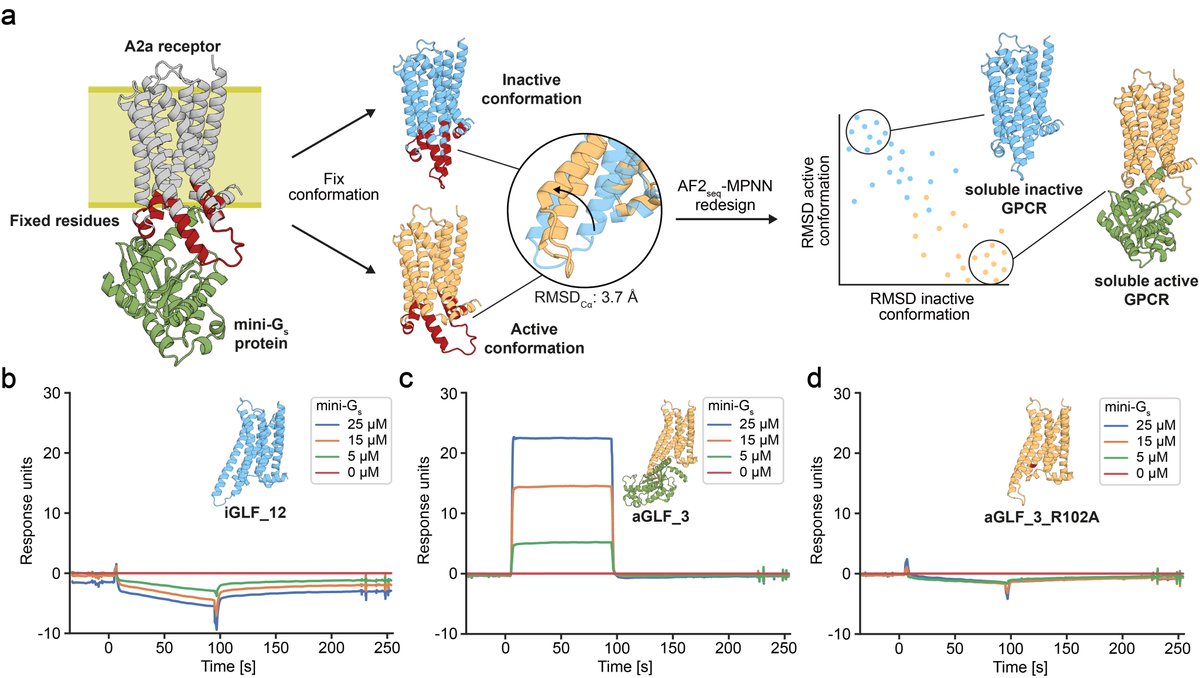
Great work by @CasperGoverde @GoldbachNico, Petra Balbi, and @Vecchiology
• • •
Missing some Tweet in this thread? You can try to
force a refresh