Imagine that you were to experience a thunderstorm for the first time as an adult.
The sky turns dark in the middle of the day and lightning bolts start coming out of the sky.
This couldn’t seem natural.
This is kind of how I see things with COVID origins debates.
1/
The sky turns dark in the middle of the day and lightning bolts start coming out of the sky.
This couldn’t seem natural.
This is kind of how I see things with COVID origins debates.
1/

I’m from the Midwest, so tornadoes and violent thunderstorm seem normal to me.
I’m also a virologist, so some of the wild stuff that Coronaviruses do seems normal too.
2/
I’m also a virologist, so some of the wild stuff that Coronaviruses do seems normal too.
2/
No one is 100% sure where SARS-CoV-2 came from, so I won’t argue that it’s natural.
I’m just going to explain (again) why a natural origin would not be surprising at all to me.
I’m not calling anyone stupid, I’m just explaining my point of view.
3/
I’m just going to explain (again) why a natural origin would not be surprising at all to me.
I’m not calling anyone stupid, I’m just explaining my point of view.
3/
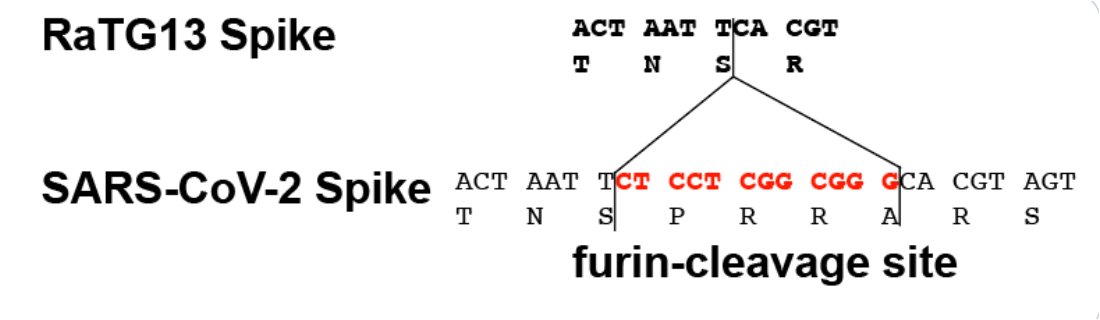
The biggest difference between SARS-CoV-2 and the other Sarbecoviruses is the furin-cleavage site (FCS) that was inserted between S1 and S2, which has been argued is proof the virus was engineered.
There are other arguments too, but the FCS is the only point I’m discussing.
4/
There are other arguments too, but the FCS is the only point I’m discussing.
4/
I’ll make 4 points.
1. Random insertions in Coronaviruses are common.
2. Insertions most often occur in genetic ‘soft spots’
3. The S1/S2 border is a clear ‘soft spot’.
4. Furin-cleave sites (FCS) are easy to make.
5/
1. Random insertions in Coronaviruses are common.
2. Insertions most often occur in genetic ‘soft spots’
3. The S1/S2 border is a clear ‘soft spot’.
4. Furin-cleave sites (FCS) are easy to make.
5/
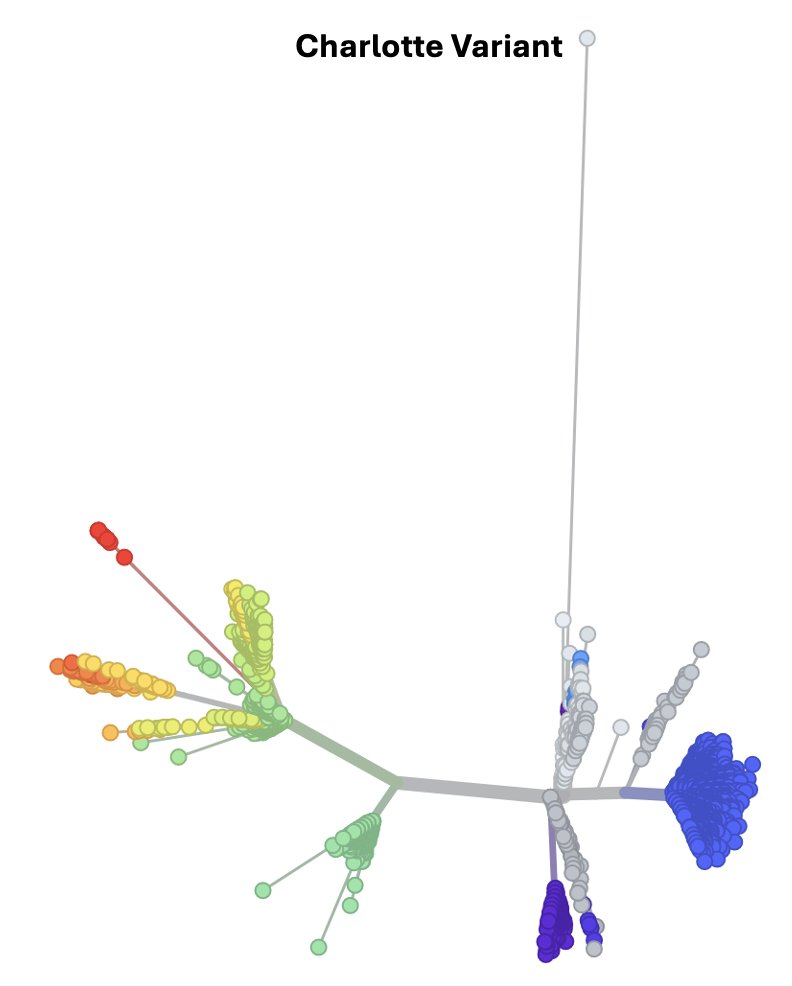
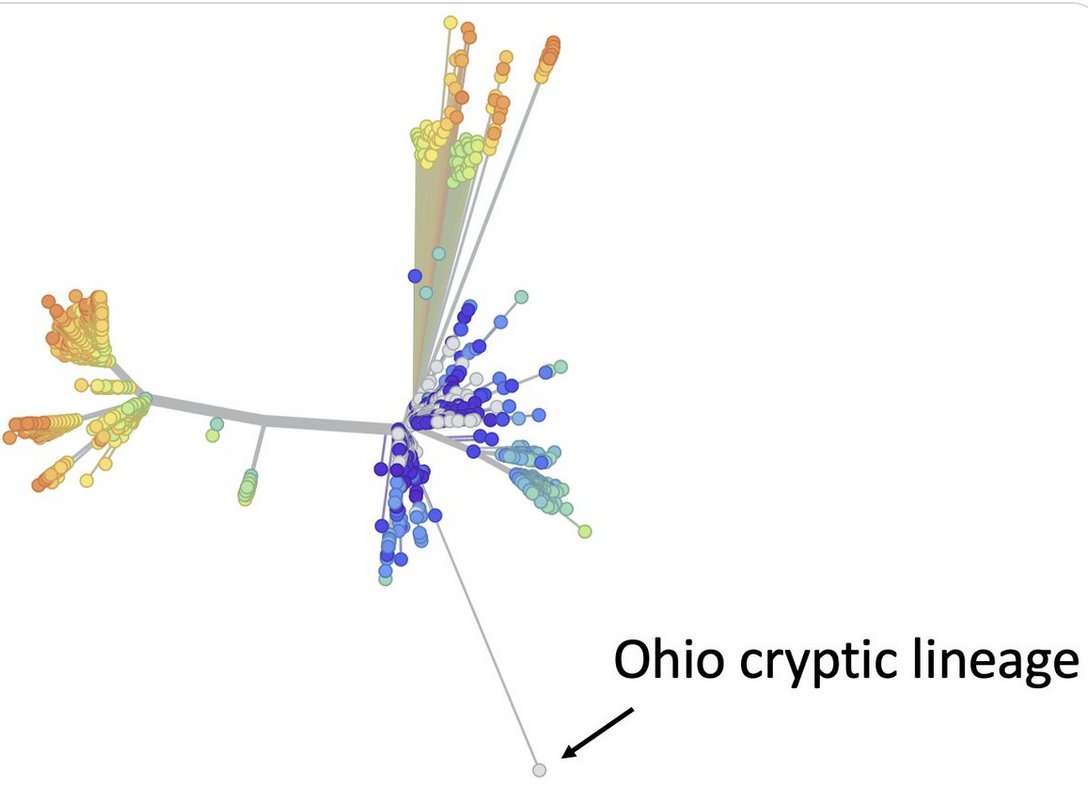
I study cryptic lineages, which appear to be SC2 persistent infections that have lasted a very long time.
We don’t know who they come from, but we detect them in wastewater.
6/doi.org/10.1371/journa…
We don’t know who they come from, but we detect them in wastewater.
6/doi.org/10.1371/journa…
We’ve been working on a project to curate the whole genome of as many cryptic lineages as possible from wastewater sequences deposited into public databases.
We’ve curated 18 so far and they are pretty diverse.
7/
We’ve curated 18 so far and they are pretty diverse.
7/
One of the first things we noticed about these lineages is that insertions are really common. About a third of the cryptic lineages had at least one insertion.
The insertions are not random. There are ‘soft spots’ where insertions are tolerated.
8/
The insertions are not random. There are ‘soft spots’ where insertions are tolerated.
8/
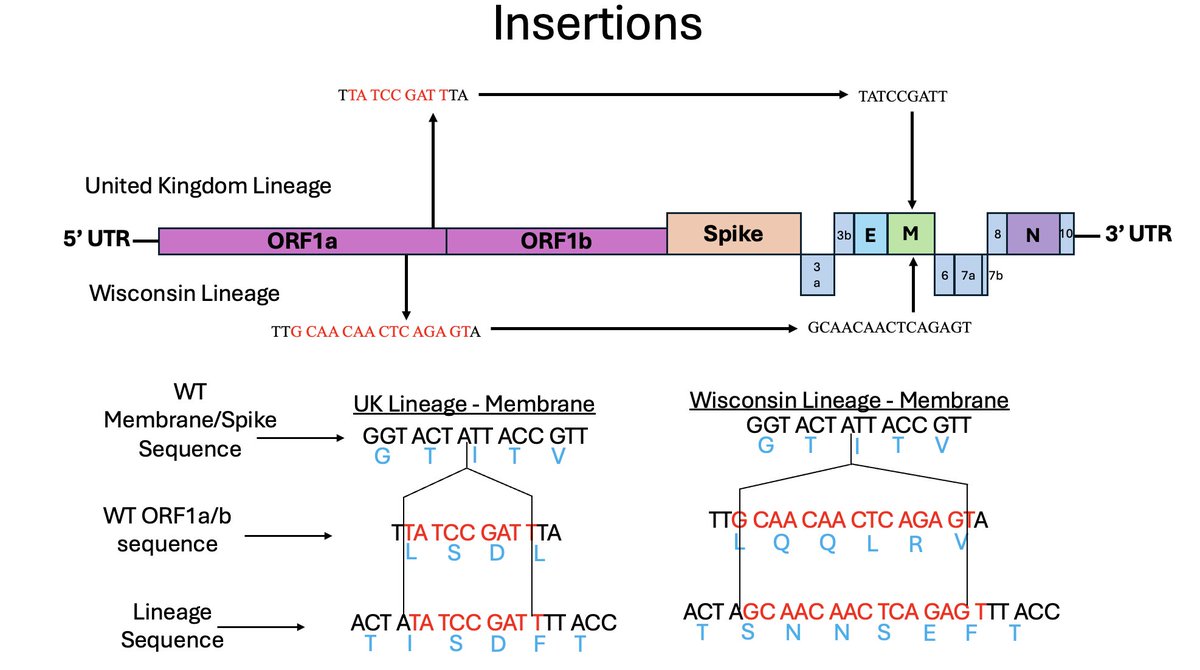
Here is an example.
2 of the 18 cryptic sequences contained an insertion in *precisely* the same site in the M gene.
Both were duplications from Orf1a, but they were completely different insertions.
9/
2 of the 18 cryptic sequences contained an insertion in *precisely* the same site in the M gene.
Both were duplications from Orf1a, but they were completely different insertions.
9/
Other soft spots in the SARS-CoV-2 genome are in the NTD of Spike, and at the S1/S2 border.
We know because we’ve seen lots of insertions occur there.
Here’s an example of an S1/S2 insert in the lineage AT.1, which circulated in 2021.
10/
We know because we’ve seen lots of insertions occur there.
Here’s an example of an S1/S2 insert in the lineage AT.1, which circulated in 2021.
10/
https://twitter.com/PeacockFlu/status/1447578684664254471
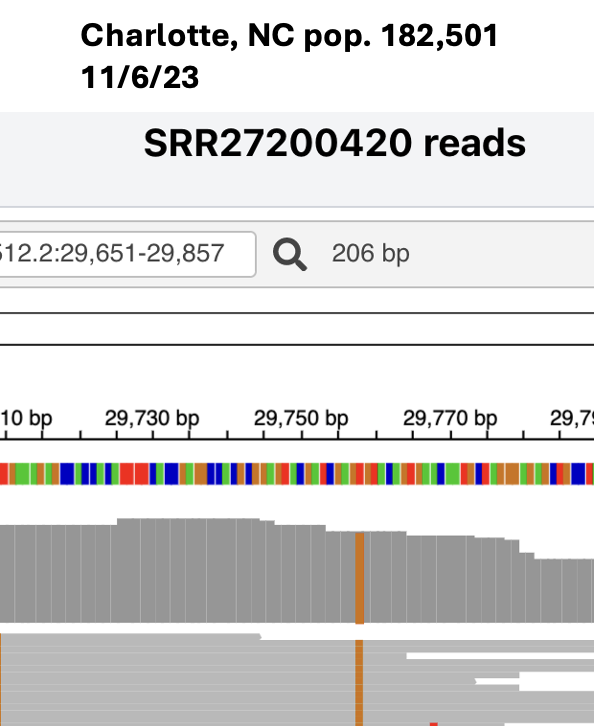
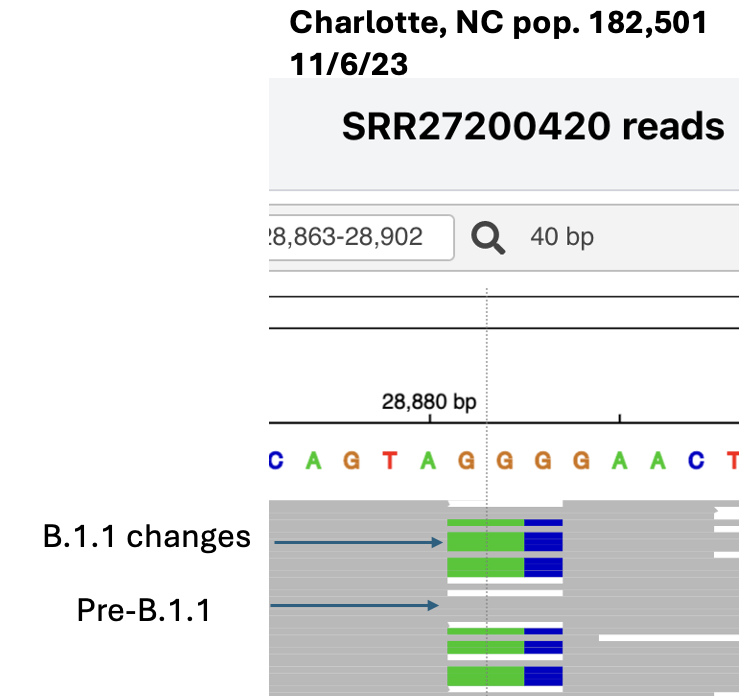
Here’s another example, an insertion just before the FCS in a BA.1.1 sub-lineage that circulated in early 2022.
In this case we don't know for sure where the insertion sequence came from, probably a human RNA.
11/
In this case we don't know for sure where the insertion sequence came from, probably a human RNA.
11/
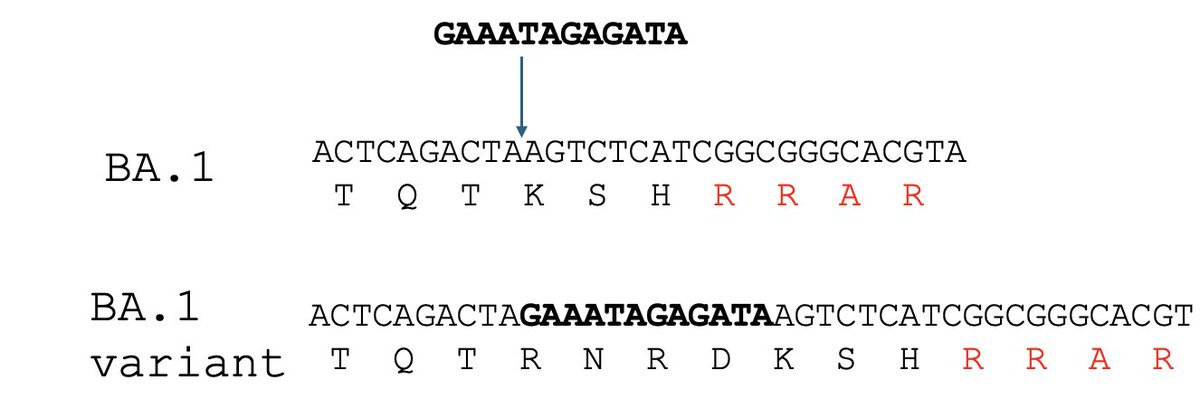
So insertions are common, and the place where the furin-cleave site (FCS) in SC2 appeared is a common place to see them.
So what is a furin-cleavage site?
12/
So what is a furin-cleavage site?
12/
A furin-cleavage site is a loosely defined 4 AA sequence that the cellular protease furin can cut.
We can try to predict what will be a good FCS, but ultimately the protease gets to decide.
13/
We can try to predict what will be a good FCS, but ultimately the protease gets to decide.
13/
We’ve seen very diverse sequences that furin will cut.
They always have at least one arginine (R), but more often two or three.
14/
They always have at least one arginine (R), but more often two or three.
14/
As it happens, R is one of the most common codons.
6/64 possible codons produce R.
If you typed the sequence to produce 4 codons at random (12 nt), there is a 32.5% chance that at least one of them is going to be an Arg (R).
15/
6/64 possible codons produce R.
If you typed the sequence to produce 4 codons at random (12 nt), there is a 32.5% chance that at least one of them is going to be an Arg (R).
15/
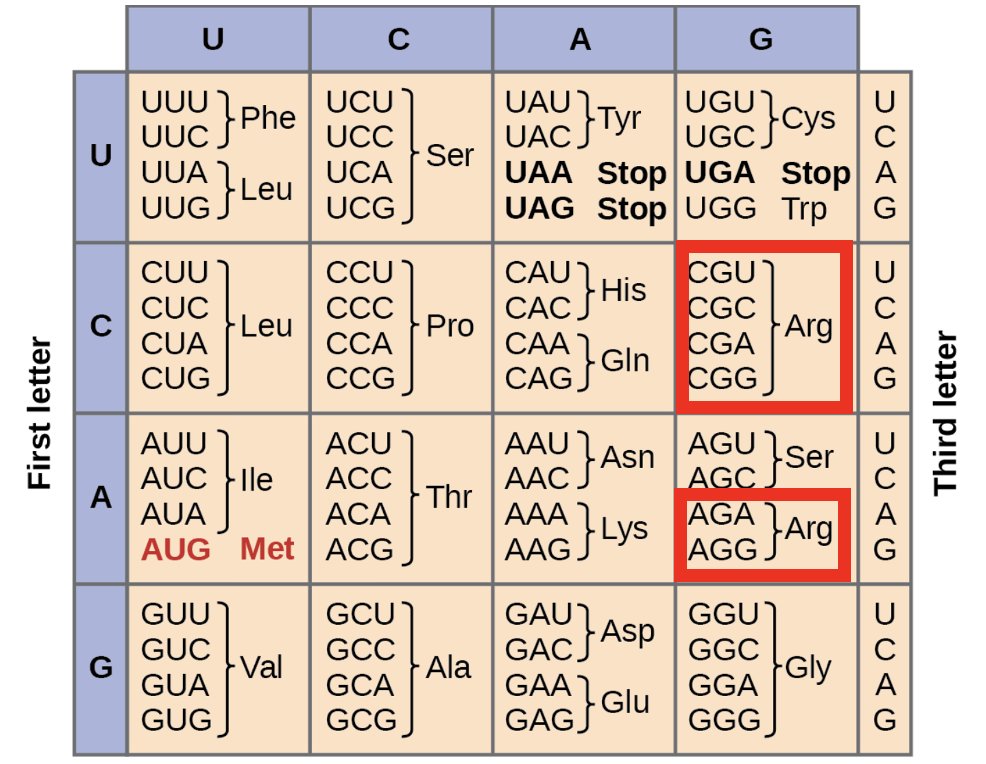
There are over 17,000 different sequences (~0.1% of possible combinations) that produce the 'canonical' FCS (R-X-(K/R)-R).
There are also many more 'non-canonical' sequences that still work.
16/
There are also many more 'non-canonical' sequences that still work.
16/
It’s obvious that having an FCS at the S1/S2 border can be advantageous to the virus in certain situations because we see it there a lot.
That’s why investigators (DEFUSE) had proposed to study this long before SC2 existed.
17/
sciencedirect.com/science/articl…
That’s why investigators (DEFUSE) had proposed to study this long before SC2 existed.
17/
sciencedirect.com/science/articl…
People will ask if I’ve seen an FCS acquired in real time, or if I've see one acquired by a Sarbecovirus (which is pretty specific).
This is sort of like asking ‘have you seen a lightning strike that looks exactly like this?’.
18/
This is sort of like asking ‘have you seen a lightning strike that looks exactly like this?’.
18/
The answer is No, I have not seen precisely that, but that doesn’t mean it hasn’t happened, or that it is surprising.
I see much stranger things all the time.
Shit happens.
19/
I see much stranger things all the time.
Shit happens.
19/
• • •
Missing some Tweet in this thread? You can try to
force a refresh