𝘼 𝙈𝘼𝙎𝙏𝙀𝙍𝙋𝙄𝙀𝘾𝙀 𝙖𝙗𝙤𝙪𝙩 𝙩𝙝𝙚 𝙀𝙑𝙊𝙇𝙐𝙏𝙄𝙊𝙉 𝙤𝙛 𝙎𝘼𝙍𝙎-𝘾𝙊𝙑-2
I have already posted an extract from this study, which I consider to be 𝙤𝙣𝙚 𝙤𝙛 𝙩𝙝𝙚 𝙢𝙤𝙨𝙩 𝙞𝙢𝙥𝙤𝙧𝙩𝙖𝙣𝙩 𝙨𝙩𝙪𝙙𝙞𝙚𝙨 I have read. Having been able to obtain the complete PDF,
I have already posted an extract from this study, which I consider to be 𝙤𝙣𝙚 𝙤𝙛 𝙩𝙝𝙚 𝙢𝙤𝙨𝙩 𝙞𝙢𝙥𝙤𝙧𝙩𝙖𝙣𝙩 𝙨𝙩𝙪𝙙𝙞𝙚𝙨 I have read. Having been able to obtain the complete PDF,

2) I would like to come back to it, in a very very long thread, in 2 parts:
▶️ 𝙖 𝙗𝙧𝙞𝙚𝙛 𝙎𝙪𝙢𝙢𝙖𝙧𝙮
▶️ 𝙖 𝙢𝙤𝙧𝙚 𝙙𝙚𝙩𝙖𝙞𝙡𝙚𝙙 𝙀𝙭𝙥𝙡𝙖𝙣𝙖𝙩𝙞𝙤𝙣 𝙬𝙞𝙩𝙝 𝙜𝙧𝙖𝙥𝙝𝙞𝙘𝙨
▶️ 𝙖 𝙗𝙧𝙞𝙚𝙛 𝙎𝙪𝙢𝙢𝙖𝙧𝙮
▶️ 𝙖 𝙢𝙤𝙧𝙚 𝙙𝙚𝙩𝙖𝙞𝙡𝙚𝙙 𝙀𝙭𝙥𝙡𝙖𝙣𝙖𝙩𝙞𝙤𝙣 𝙬𝙞𝙩𝙝 𝙜𝙧𝙖𝙥𝙝𝙞𝙘𝙨

3) 𝗕𝗥𝗜𝗘𝗙 𝗦𝗨𝗠𝗠𝗔𝗥𝗬
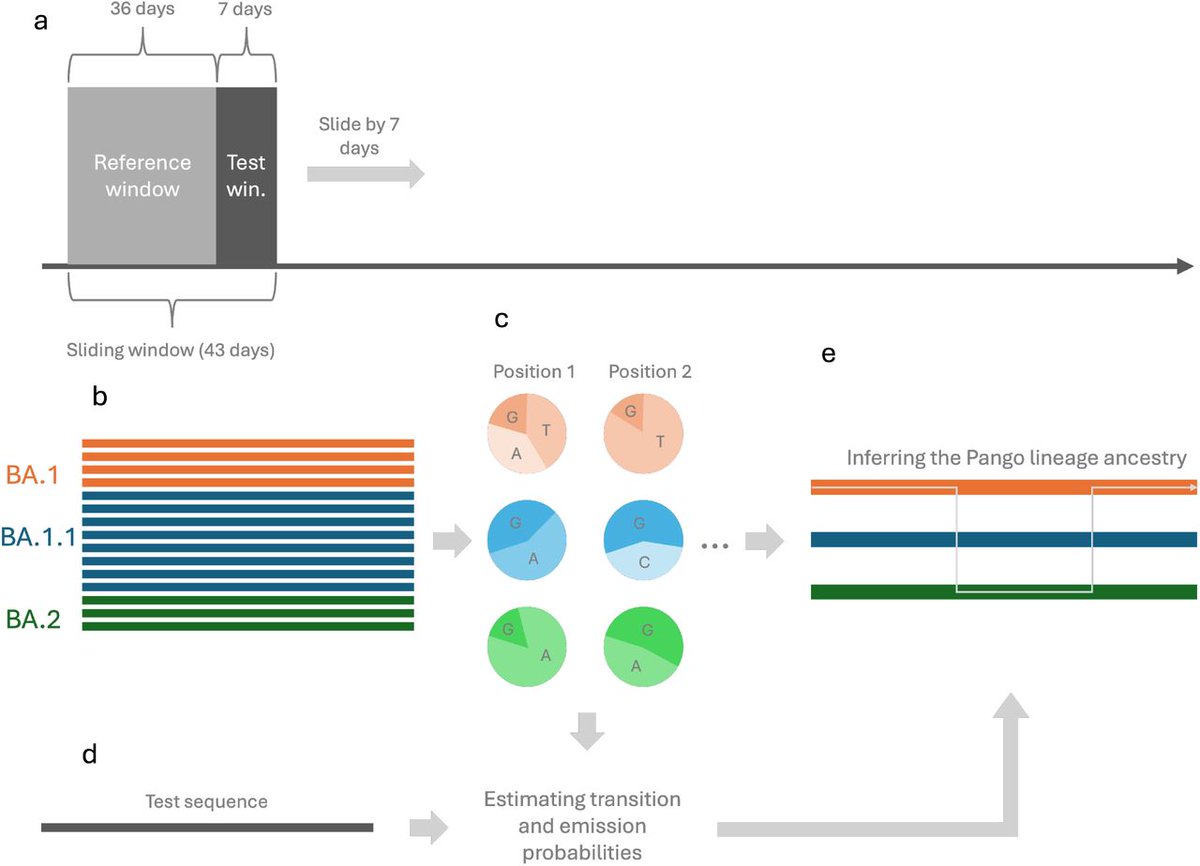
▶️ The authors conducted a comprehensive analysis of over 141,000 unique missense mutations detected in SARS-CoV-2 proteins within the first 2.5 years of the pandemic, based on data from the GISAID database.
▶️ The authors conducted a comprehensive analysis of over 141,000 unique missense mutations detected in SARS-CoV-2 proteins within the first 2.5 years of the pandemic, based on data from the GISAID database.

4) ▶️ They found that by October 2022, over 98% of all possible conservative amino acid substitutions had occurred. Non-conservative mutations were also highly represented at nearly 74%.
▶️ For many residues, all 19 possible amino acid replacements were observed,
▶️ For many residues, all 19 possible amino acid replacements were observed,
5) ... suggesting the virus explored all mutation possibilities within proteins.
▶️ There was a plateau in new mutations detected after July 2022, indicating mutations not observed likely impair virus survival.
▶️ Mutation levels varied between proteins ...
▶️ There was a plateau in new mutations detected after July 2022, indicating mutations not observed likely impair virus survival.
▶️ Mutation levels varied between proteins ...

6) ▶️ ... but most sites showed high variability, with an average of 14-16 substitutions per site.
▶️ Terminal residues were generally more conserved due to protease cleavage. Structured/ disordered regions showed similar mutation levels.
▶️ Terminal residues were generally more conserved due to protease cleavage. Structured/ disordered regions showed similar mutation levels.
7) ▶️ Rarer mutations involved bulkier replacements, while small hydrophobic residues were more tolerant of changes.
▶️ Highly mutated proteins included Spike and NSP3, while NSP9, E and NSP7 displayed relative conservation.
▶️ Highly mutated proteins included Spike and NSP3, while NSP9, E and NSP7 displayed relative conservation.

8) ▶️ The detection of such a huge amount of single-point mutations demonstrates that variants currently present worldwide are the results of a very stringent selection process that has taken place during the pandemic.
9) ▶️ Indeed, randomly selected sequences of the currently most diffuse (Dec. 2023) SARS-CoV-2 variants (JN.1 and HV.1) contains only 84 and 75 single point amino acid mutations compared to the first sequenced genome of the virus (Wuhan genome)
10) ▶️ In summary, the study amassed a large dataset characterizing SARS-CoV-2 protein evolution through mutations, finding the virus explored almost all possible substitutions in the first 2.5 years of the pandemic.
Thanks for reading 🙏
(2𝘯𝘥 𝘱𝘢𝘳𝘵, 𝘷𝘦𝘳𝘺 𝘴𝘰𝘰𝘯)
Thanks for reading 🙏
(2𝘯𝘥 𝘱𝘢𝘳𝘵, 𝘷𝘦𝘳𝘺 𝘴𝘰𝘰𝘯)
11) 𝗦𝗢𝗠𝗘 𝗘𝗫𝗣𝗟𝗔𝗡𝗔𝗧𝗜𝗢𝗡𝗦
This study discusses the challenges of understanding how viruses adapt to host organisms and the mechanisms they use for information transmission. The ongoing COVID-19 pandemic has provided a wealth of information about the SARS-CoV-2 virus
This study discusses the challenges of understanding how viruses adapt to host organisms and the mechanisms they use for information transmission. The ongoing COVID-19 pandemic has provided a wealth of information about the SARS-CoV-2 virus
12) ... and its variants. In this study, they focuse on single point mutations (small changes or alterations in the RNA sequence) detected during the first years of the pandemic and reveals that the virus has undergone progressive adaptation to the host, 

13) ... resulting in an accumulation of hydrophobic residues. They collected and classified amino acid mutations in the virus's proteins, focusing on missense mutations that are tolerated by the virus and do not compromise its vitality. The mutations were classified 

13) ... as conservative or non-conservative based on evolutionary acceptance probability and the similarity of encoding codons.
It is observed that there was a rapid increase in mutations in the early stage of the pandemic, but the rate of increase has since slowed down.
It is observed that there was a rapid increase in mutations in the early stage of the pandemic, but the rate of increase has since slowed down.

14) This plateau may indicate that unobserved mutations could be highly detrimental to the structure and function of the proteins. The time evolution of mutations suggests that there is a balance between conservative and non-conservative mutations. 

15) The analysis also highlights that most of the sites in the virus proteins have a tendency to mutate, with an average of 14.6 substitutions per site. However, there are a small number of sites that are hyper-conserved and located at the termini of the polypeptide chains. 

16) The study discusses also the impact of terminal residues and disordered regions on the mutation rate of proteins. It states that terminal residues, which are generally disordered, tend to lower the substitution level of residues.
17) When the first and last 10 residues are omitted from the analysis, there is a difference in the average number of substitutions for different amino acids. Residues in disordered regions have a higher substitution degree. Rare replacements involve pairs of residues ... 

17) with different sizes and polarity. The replacement of aspartic acid and asparagine with tryptophan has the lowest frequencies. Most replacements involving tryptophan have frequencies lower than 0.4. It highlights the asymmetry of the replacement matrix, ... 

18) ...where some substitutions occur more frequently in one direction than the reverse.
The analysis of individual SARS-CoV-2 proteins shows a wide range of substitution frequencies, with non-structural proteins having lower substitution levels compared to structural proteins.
The analysis of individual SARS-CoV-2 proteins shows a wide range of substitution frequencies, with non-structural proteins having lower substitution levels compared to structural proteins.

19) The analysis reveals that the proteins NSP9 and E have the lowest levels of substitution. NSP9 is a dimeric protein that plays a crucial role in viral replication and RNA reproduction. The regions of NSP9 with the lowest substitution values are involved 

20) .. in important structural and functional roles. The protein E is the second least mutated protein in the virus and is involved in virus particle production and maturation. Protein E has rare residues in the N-terminal region and shows variability in the C-terminal region. 

21) Protein M is involved in virus assembly and has a high degree of variability.
The Spike glycoprotein, responsible for virus entry, shows the highest variability among all the proteins. Specific residues in the Spike protein have been linked to genetic variants of concern.
The Spike glycoprotein, responsible for virus entry, shows the highest variability among all the proteins. Specific residues in the Spike protein have been linked to genetic variants of concern.

22) These residues also have a high degree of substitutions, indicating multiple attempts by the virus to mutate. The analysis also shows that the virus proteins are extremely tolerant to single-point mutations, with nearly 39% of residues being substituted ...
23) with all other 19 amino acid alternatives. Overall, the findings indicate that the currently present variants of the virus are the result of a stringent selection process during the pandemic. 

24) 𝗣𝗘𝗥𝗦𝗢𝗡𝗔𝗟 𝗢𝗣𝗜𝗡𝗜𝗢𝗡
In this study, the % of the detected substitutions compared to the possible theoretical ones was 83.6% for the Spike. It means that the Spike has still 16.4% possible substitutions. We saw, how the mutation L455S (JN.1*)
In this study, the % of the detected substitutions compared to the possible theoretical ones was 83.6% for the Spike. It means that the Spike has still 16.4% possible substitutions. We saw, how the mutation L455S (JN.1*)
https://twitter.com/ejustin46/status/1748588677532443005?t=i_Li7Hj1fNShO2O8qE-uLQ&s=19
25) .. has changed the evolution of the pandemic. For the E protein, the % of substitution was 60.4%. It is true that it is one of the most stable but we saw, with the mutation T9I, hallmark of the Omicron, its impact ...
https://twitter.com/ejustin46/status/1705595823587758560?t=i_Li7Hj1fNShO2O8qE-uLQ&s=19
26) ... in term of pathogenicity.
New mutations with the spike, envelope, or others proteins can still radically change the virus.
Their emergence will perhaps take longer, but in our opinion we are far from the end of history.
Thanks for reading 🙏
New mutations with the spike, envelope, or others proteins can still radically change the virus.
Their emergence will perhaps take longer, but in our opinion we are far from the end of history.
Thanks for reading 🙏
Thanks to Lee @AltenbergLee for the link
https://twitter.com/AltenbergLee/status/1772993517444354191?t=nVbkThpA0_VDYRr9VLoa-g&s=19
• • •
Missing some Tweet in this thread? You can try to
force a refresh